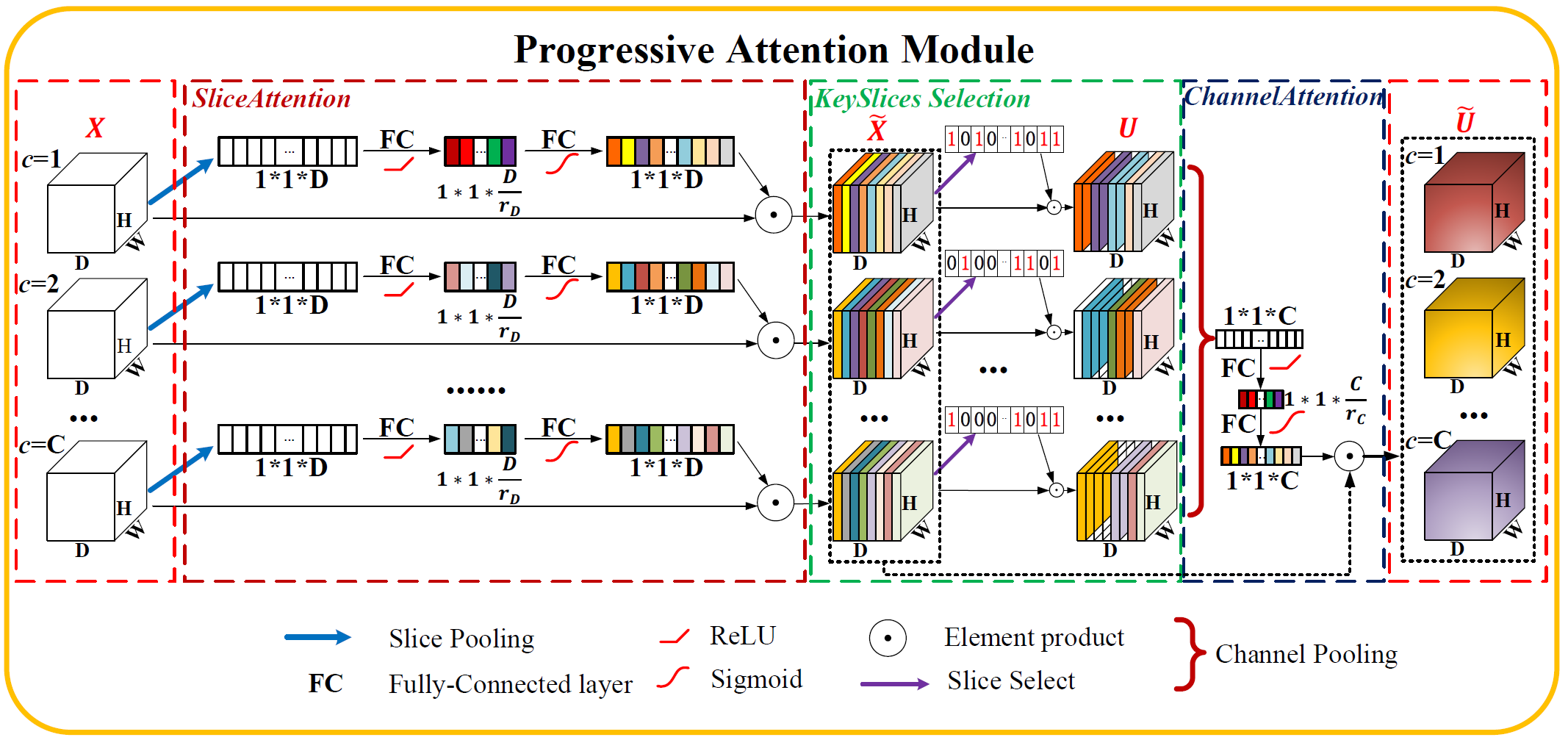
Progressive Attention Module for Segmentation of Volumetric Medical Images
By Minghui Zhang, Hong Pan, Yaping Zhu, and Yun Gu
Institute of Medical Robotics, Shanghai Jiao Tong University
Department of Computer Science and Software Engineering, Swinburne University of Technology
Medical Image Segmentation Based on Attention mechanism(Fine-Tune for natural image[semantic/instance] segmentation).
This project is dedicated to
- Collecting and re-implementing basic models and different attention mechanisms, transforming them modular and portable.
- Proposing a novel attention mechanism ———— 'Slice Attention' and 'Progressive Attention Module' tailored for 3D data Segmentation.
Main purpose is used in 3D Medical Image Segmentation. Fine-tune for Other CV tasks need attention is easily meanwhile.
We have used three medical image dataset,both 3D MRI format.You can use other formats like CT only need to constrcuct a concrete datatset loader
in /data.
| Dataset | Link |
|---|---|
| BraTS 2018 dataset | link |
| MALC dataset | link |
| HVSMR dataset | link |
This section include basic model(for segmentation or feature extraction) and different attention mechanisms.Each attention mechanism can recalibrate multi-dim feature maps across their own functional domain.
Most attention mechanisms can be modularized and integrated into any sub feature maps(e.g. each encoder in 3D UNet or each block in VNet) if not special noted.
All models and basic modules are in /models,We provide the pre-trained Models in Model_zoo.md:
Here,we integrate and provide brief description for basic segmentation models and attention modules used in our experiments. Most of them have conducted and you can find pretrained models in Model_zoo.md. Some modules have not finished and we will keep conducting experiments and updating this project.
| Module | Paper | Name |
|---|---|---|
| 3D UNet | link | 3D UNet |
| VNet | link | VNet |
| U-Net | link | 2D UNet |
| DeepMedic | link | DeepMedic |
| VoxResNet | link | VoxResNet |
| H-DenseUNet | link | H-DenseUNet |
| Module | Paper | Name |
|---|---|---|
| Squeeze-and-Excitation | link | SE |
| Convolutional Block Attention Module | link | CBAM |
| Project&Excitation | link | PENet |
| Attention U-Net | link | AG |
| AnatomyNet | link | AnatomyNet |
| Progressive Attention Module | link | PANet |
| Class Activation Map | link | CAM |
| Spatial Transformer Net | link | STN |
| Split Attention | link | SpA |
To fully understand the architecture of our projects, please take a reference to
The Technical Report has clearly demonstrate the procedure of training and test, including loading data, pre-processing data, loading pretrained models, training and testing models, saving model weights and other log info, etc.
You can use the following command to train a model, demo_train.py is a script with detailed annotations.
python demo_train.py --dataroot $DATASET_DIR \
--name $EXPERIMENT_NAME \
--checkpoints_dir $MODEL_SAVEDIR \
--model $MODEL \
--dataset_mode $DATASET_MODE \
--in_channels $INPUT_CH \
--out_channels $OUTPUT_CH \
--gpu_ids $GPU_IDS
You can use the following command to test a model, demo_test.py is a script with detailed annotations.
python demo_test.py --dataroot $DATASET_DIR \
--name $EXPERIMENT_NAME \
--checkpoints_dir $MODEL_LOADDIR \
--model $MODEL \
--dataset_mode $DATASET_MODE \
--in_channels $INPUT_CH \
--out_channels $OUTPUT_CH \
--gpu_ids $GPU_IDS
The research in this study is carried out on both Windows10 and Ubuntu16.04 with 4 NVIDIA 1080Ti GPU with 44GB memory.
The deep learning framework is PyTorch≥1.1.0 and Torchvision ≥0.4.0.
Some python libraries are also necessary, you can use the following command to set up.
pip install -r requirements.txt
If you find this repository or our paper useful, please consider citing our paper:
@article{zhang2022progressive,
title={Progressive attention module for segmentation of volumetric medical images},
author={Zhang, Minghui and Pan, Hong and Zhu, Yaping and Gu, Yun},
journal={Medical Physics},
volume={49},
number={1},
pages={295--308},
year={2022},
publisher={Wiley Online Library}
}