Code for the paper Ament-Velásquez et al. (2020) Mycokeys 75: 51-69.
In this study we compiled sequence data for a number of molecular markers (ITS, LSU, rpb2, and beta-tubulin) to resolve the phylogenetic relationships of lineages within the Podosporaceae family, sensu Wang et al. (2019) Studies in Mycology 93:155-252. The primary objective was to determine the relative position of the model species Podospora anserina and the type species of the genus, Podospora fimiseda. Unfortunately, there is a lot of missing data.
The data required for this pipeline is:
- The master concatenated alignment in nexus format with annotated limits of the markers (
Podosporaceae_20200714.nxs). You can open it with SeaView, for example. We submitted it to TreeBase (accession number XXXX) but I had to fit it to the Mesquite nexus format for that. To make the Mesquite file compatible with this pipeline, you can open it in SeaView and save it as a new nexus file. It should work then. In any case, I put the original nexus here in this repo just in case. - A partition file (in the RAxML style) with the genes to be analyzed by the scripts of Shen et al. (2017) to obtain gene-wise log-likelihood scores (dGLS) values:
allmarkers_combining_orders.txt.
The scripts needed:
- The Snakemake pipeline
Podosporaceae.smk - The configuration file of the pipeline
Podosporaceae_config.yaml 1_sitewise_analyzer.pland2_genewise_analyzer.plfrom Shen et al. (2017)- An R script for plotting:
Shen2017_podofam.R
The configuration file contains the paths to the necessary files to run the pipeline.
$ cat Podosporaceae_config.yaml
# Configuration file of the Podosporaceae.smk pipeline
## Master nexus file
masternex: "data/Podosporaceae_20200714.nxs"
## Name of the full concatenation of all markers in the master nexus file
allmarkersname: "allmarkers"
## RAxML-like partition file to calculate the SLS and GLS metrics (It must be named "{allmarkersname}_combining_orders.txt")
orders: "data/allmarkers_combining_orders.txt"
## Filtering for alignments of individual markers
minfrac: 0.45 # min fraction of overlap with the whole alignment length for a sequence to be considered
minlen: 250
## Scripts
sitewise_analyzer: "scripts/1_sitewise_analyzer.pl"
genewise_analyzer: "scripts/2_genewise_analyzer.pl"
plotShen: "scripts/Shen2017_podofam.R"
# Outgroup clade
outgroup: ["Lasiosphaeria_ovina_SMH1538", "Zopfiella_tabulata_CBS230.78", "Sordaria_fimicola_SMH4106", "Diplogelasinospora_princeps_FMR13414", "Chaetomium_globosum_CBS148.51", 'Chaetomium_globosum_CBS160.62', 'Cercophora_mirabilis_CBS120402']
# A representative of clades A, B and C in that order
testmonophyly: ["Podospora_anserina_S", "Cercophora_grandiuscula_CBS120013", "Podospora_fimiseda_CBS990.96"]
To run the Snakemake pipeline, I constructed a conda environment. I assume in the following that you have conda installed already.
I named the environment LorePhylogenetics, but you can call it whatever you like :).
$ conda create -n LorePhylogenetics -c bioconda
$ conda activate LorePhylogenetics
$ conda install -c bioconda snakemake-minimal=5.4.4 biopython=1.72 mafft=7.407 iqtree=1.6.8 raxml=8.2.12
$ conda install r-tidyr=1.1.0 # included dplyr 1.0.0
$ conda install r-cowplot=1.0.0 # it comes with ggplot2 3.1.1
$ conda install -c etetoolkit ete3=3.1.1
First, to get an idea of how the pipeline looks like we can make a rulegraph:
$ conda install -c pkgs/main graphviz=2.40.1 # already there
$ snakemake --snakefile Podosporaceae.smk --configfile Podosporaceae_config.yaml --rulegraph | dot -Tpng > rulegraph.png
However, in this case I'm using Checkpoints right away so almost all of the graph won't be calculated until that checkpoint is ran.
For testing without running the pipeline:
$ conda activate LorePhylogenetics
$ snakemake --snakefile Podosporaceae.smk --configfile Podosporaceae_config.yaml -pn
To run the pipeline I like to make a screen in case the computer is turned off or something.
$ screen -R phylo
$ conda activate LorePhylogenetics
$ snakemake --snakefile Podosporaceae.smk --configfile Podosporaceae_config.yaml -p -j 40 --keep-going --use-conda &> Podosporaceae.log &
[1] 40092
Notice the use of the -j argument, where I specify the number of threads available for the pipeline.
If we produce the graph again:
$ snakemake --snakefile Podosporaceae.smk --configfile Podosporaceae_config.yaml --rulegraph | dot -Tpng > rulegraph.png
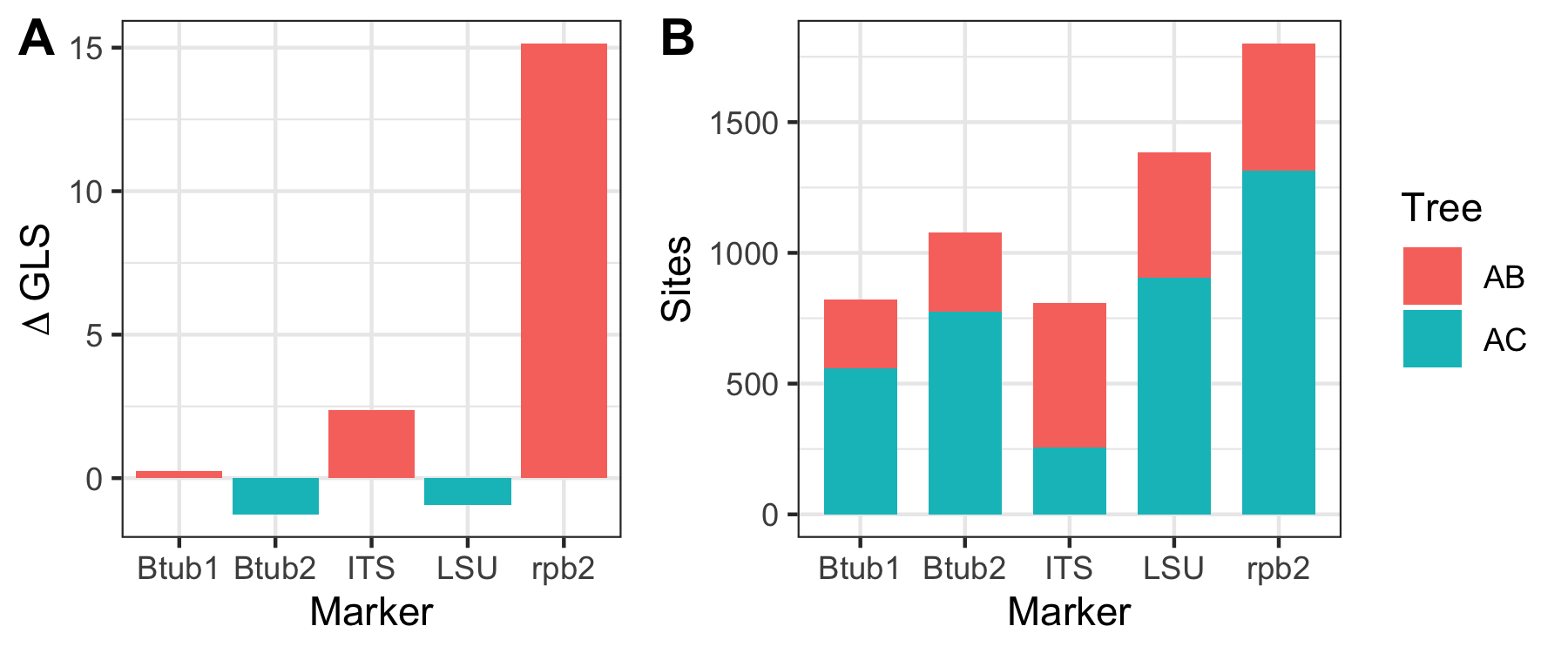
After the pipeline runs successfully, there will be a new folder called results that contains the individual ML trees of each partition in the nexus file, as well as a plot with the dGLS per marker and the number of sites supporting T1 and T2.
- ShenMetrics.pdf - Figure 4 in the paper; dGLS values per gene (A) and the number of sites supporting each topology for each gene (B) (see below)
- Tree files of all partitions (post filtering)