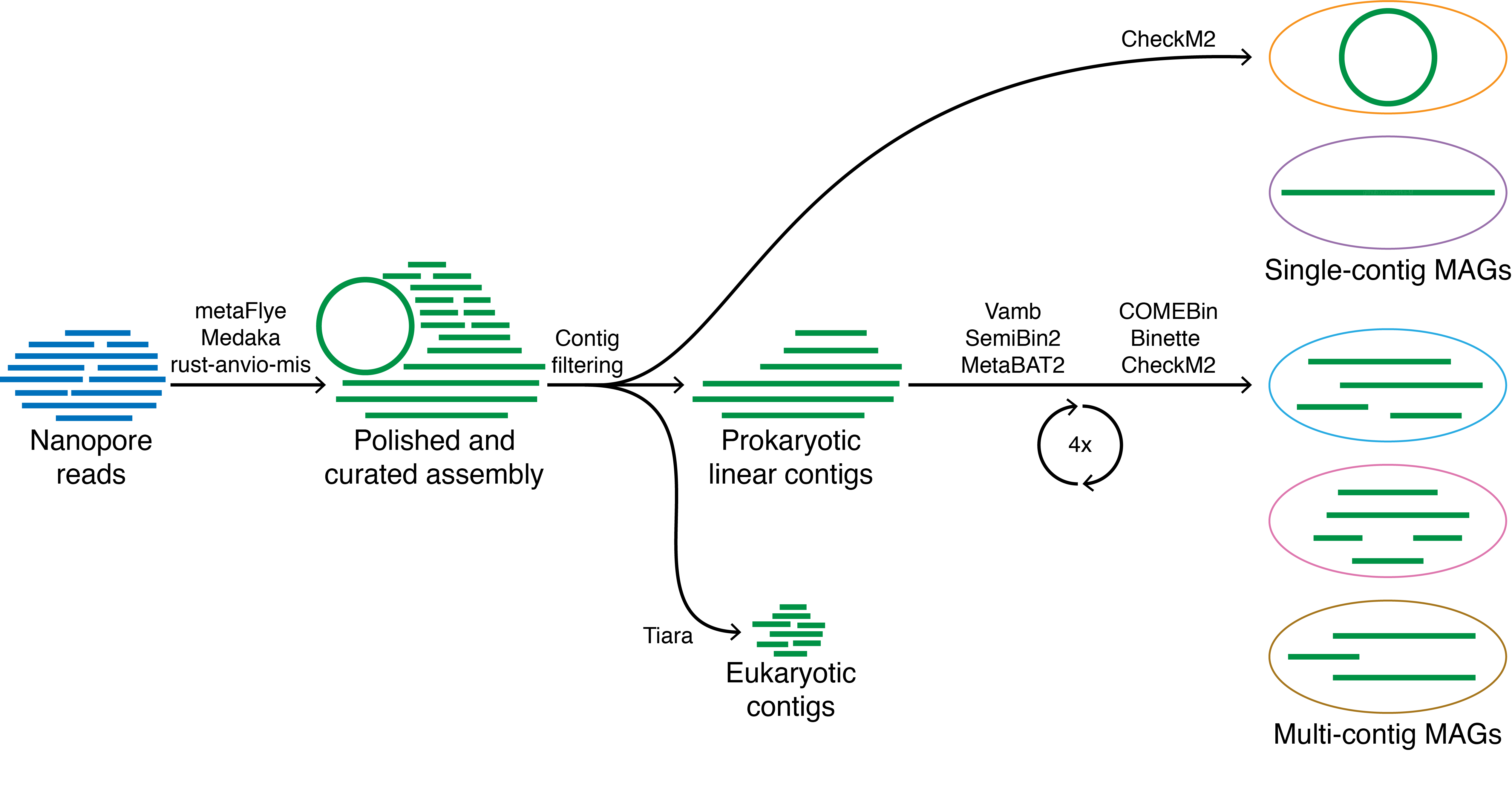
Lightweight workflow for microbial genome recovery using either Nanopore or PacBio HiFi reads.
mmlong2-lite is the microbial genome production part of the mmlong2 pipeline.
- Snakemake workflow running dependencies from a Singularity container for enhanced reproducibility
- Bioinformatics tool and parameter optimizations for high complexity metagenomics samples
- Circular microbial genome extraction as separate genome bins
- Eukaryotic contig removal for reduced microbial genome contamination
- Differential coverage support for improved microbial genome recovery
- Iterative ensemble binning strategy for improved microbial genome recovery
The mmlong2-lite workflow is available through Bioconda:
mamba install -c bioconda mmlong2-lite
To create a local Conda environment for running mmlong2-lite workflow, just copy-paste the following:
mamba create --prefix mmlong2-lite -c conda-forge -c bioconda snakemake=8.2.3 singularity=3.8.6 zenodo_get=1.6.1 pv=1.6.6 pigz=2.6 tar=1.34 -y
mamba activate ./mmlong2-lite || source activate ./mmlong2-lite
git clone https://github.com/Serka-M/mmlong2-lite/ mmlong2-lite/repo
mv mmlong2-lite/repo/src/* mmlong2-lite/bin
chmod +x mmlong2-lite/bin/mmlong2-lite
mmlong2-lite -h
After setting up the virtual environment, the required software dependencies will be automatically installed when running the workflow for the first time.
mmlong2-lite -np nanopore_reads.fastq.gz -o output_dir -p 100
MAIN SETTINGS:
-np --nanopore_reads Path to Nanopore reads (default: none)
-pb --pacbio_reads Path to PacBio HiFi reads (default: none)
-o --output_dir Output directory name (default: mmlong2)
-p --processes Number of processes/multi-threading (default: 3)
OPTIONAL SETTINGS:
-cov --coverage CSV dataframe for differential coverage binning (e.g. NP/PB/IL,/path/to/reads.fastq)
-run --run_until Run pipeline until a specified stage completes (e.g. assembly polishing filtering singletons coverage)
-tmp --temporary_dir Directory for temporary files (default: none)
-dbg --use_metamdbg Use metaMDBG for assembly of PacBio reads (default: use metaFlye)
-med --medaka_model Medaka polishing model (default: r1041_e82_400bps_sup_v5.0.0)
-mo --medaka_off Do not run Medaka polishing with Nanopore assemblies (default: use Medaka)
-vmb --use_vamb Use VAMB for binning (default: use GraphMB)
-sem --semibin_model Binning model for SemiBin (default: global)
-mlc --min_len_contig Minimum assembly contig length (default: 3000)
-mlb --min_len_bin Minimum genomic bin size (default: 250000)
-h --help Print help information
-v --version Print workflow version number
ADVANCED SETTINGS:
-fmo --flye_min_ovlp Minimum overlap between reads used by Flye assembler (default: auto)
-fmc --flye_min_cov Minimum initial contig coverage used by Flye assembler (default: 3)
-env --conda_envs_only Use conda environments instead of container (default: use container)
-n --dryrun Print summary of jobs for the Snakemake workflow
-t --touch Touch Snakemake output files
-r --rule Run specified Snakemake rule
-x --extra_inputs Extra inputs for Snakemake config file
To perform genome recovery with differential coverage, prepare a 2-column comma-separated dataframe, indicating the additional read datatype (NP for Nanopore, PB for PacBio, IL for short reads) and read file location.
Dataframe example:
PB,/path/to/your/reads/file1.fastq
NP,/path/to/your/reads/file2.fastq
IL,/path/to/your/reads/file3.fastq.gz
The prepared dataframe can be provided to the workflow through the -cov option.
<output_name>_assembly.fasta- assembled and polished metagenome<output_name>_bins.tsv- dataframe for automated binning resultsdependencies.csv- list of dependencies used and their versionsbins- directory for metagenome assembled genomes