install.packages('remotes')
remotes::install_github('alexvpickering/revigoR')
library(revigoR)
# setup python virtualenv for scraping revigo (one time only)
setup_env()
# see ?add_path_genes for example workflow starting with limma differential expression
data(go_up1)
# submit goana result to revigo web app and download results to data_dir
data_dir <- tempdir()
scrape_revigo(data_dir, go_up1)
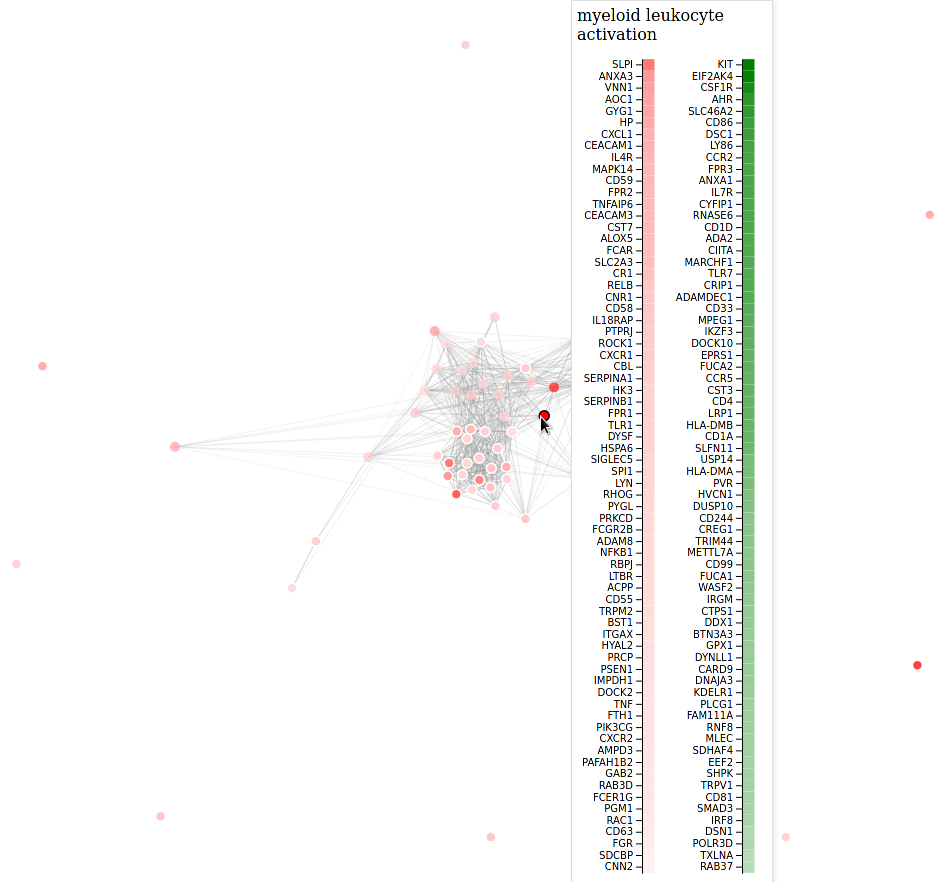
# forcegraph of revigo results (from cytoscape graph)
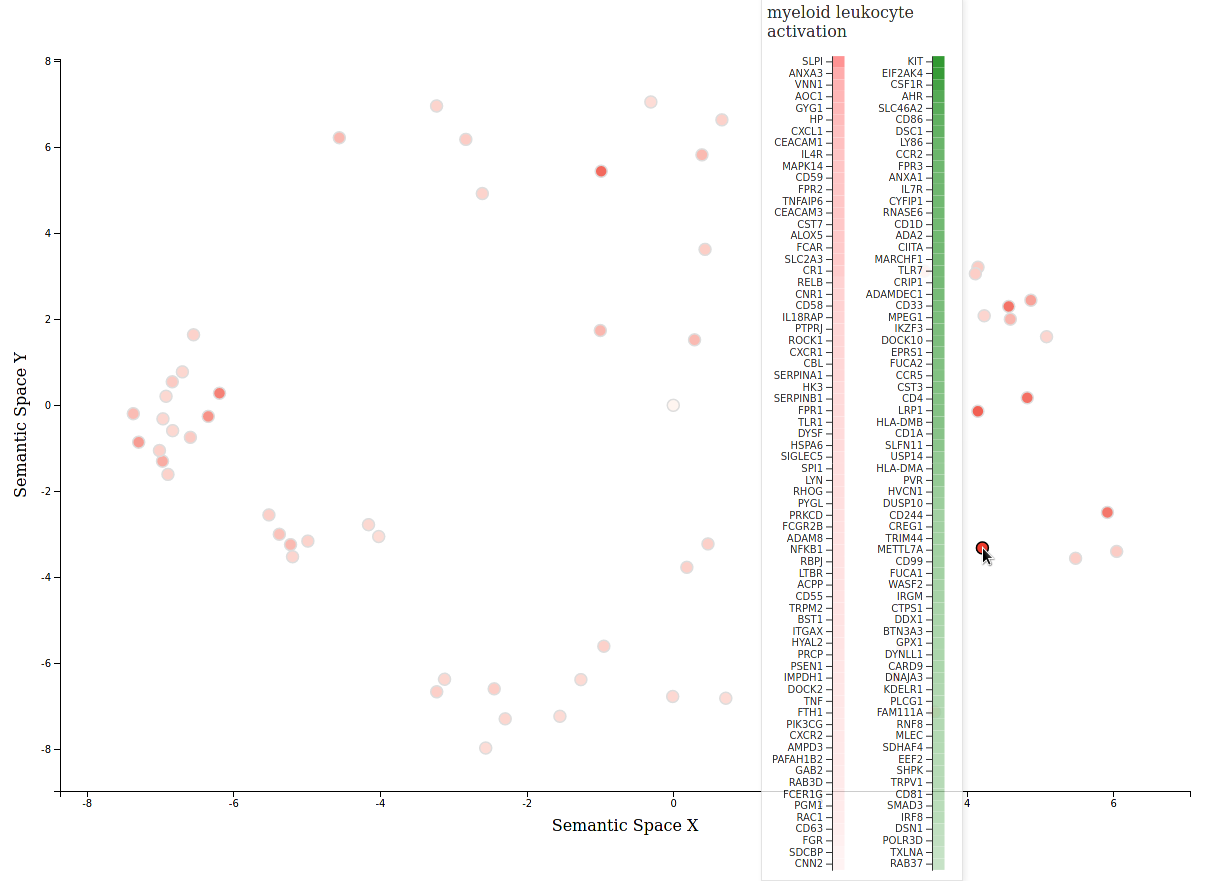
revigo_forcegraph(data_dir)An interactive scatterplot using the MDS coordinate from revigo can also be generated:
revigo_scatterplot(data_dir)Visualize where revigo merges terms across two GO analyses (shades of purple) and doesn't (shades of orange and green for each analysis respectively):
# two analyses
data(go_up2)
go_up1$analysis <- 0
go_up2$analysis <- 1
go_up <- rbind(go_up1, go_up2)
data_dir <- tempdir()
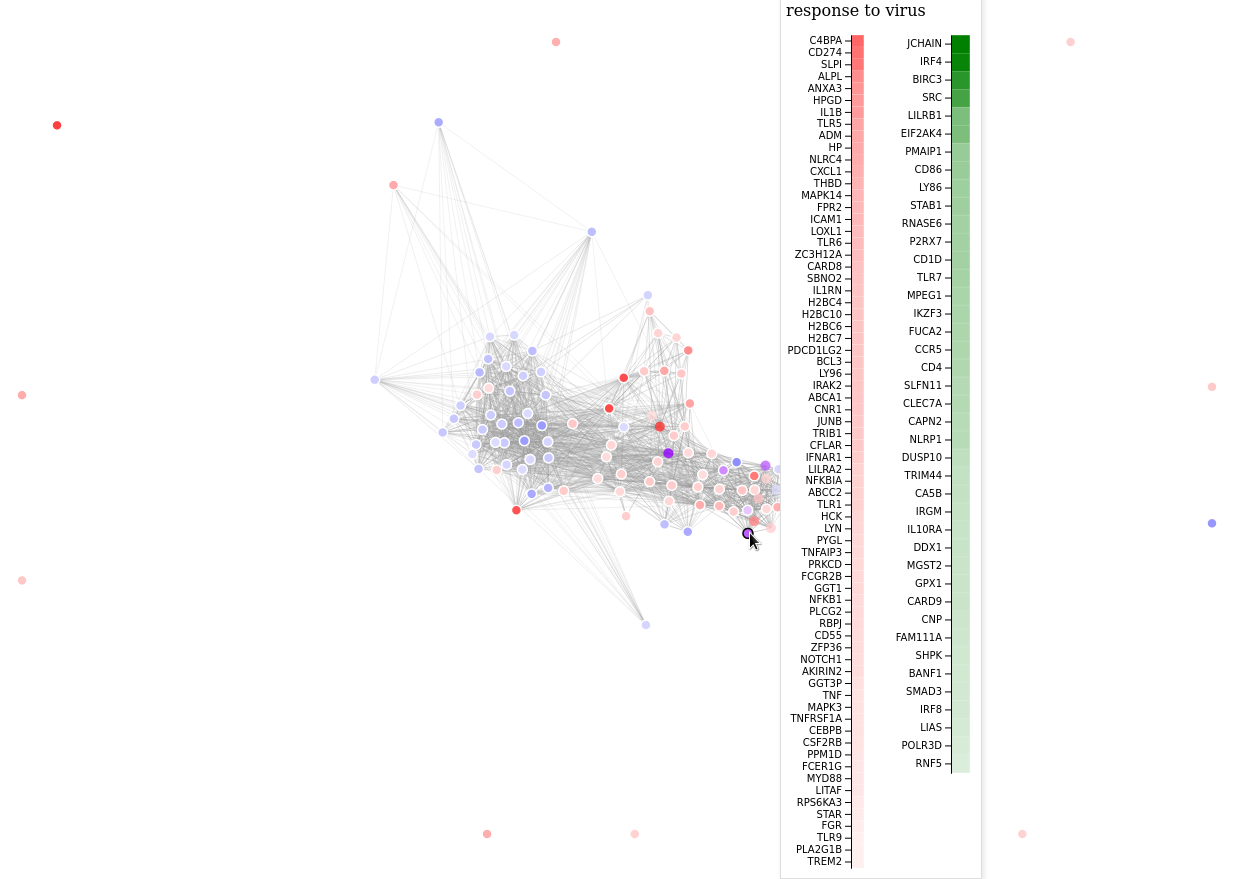
scrape_revigo(data_dir, go_up)
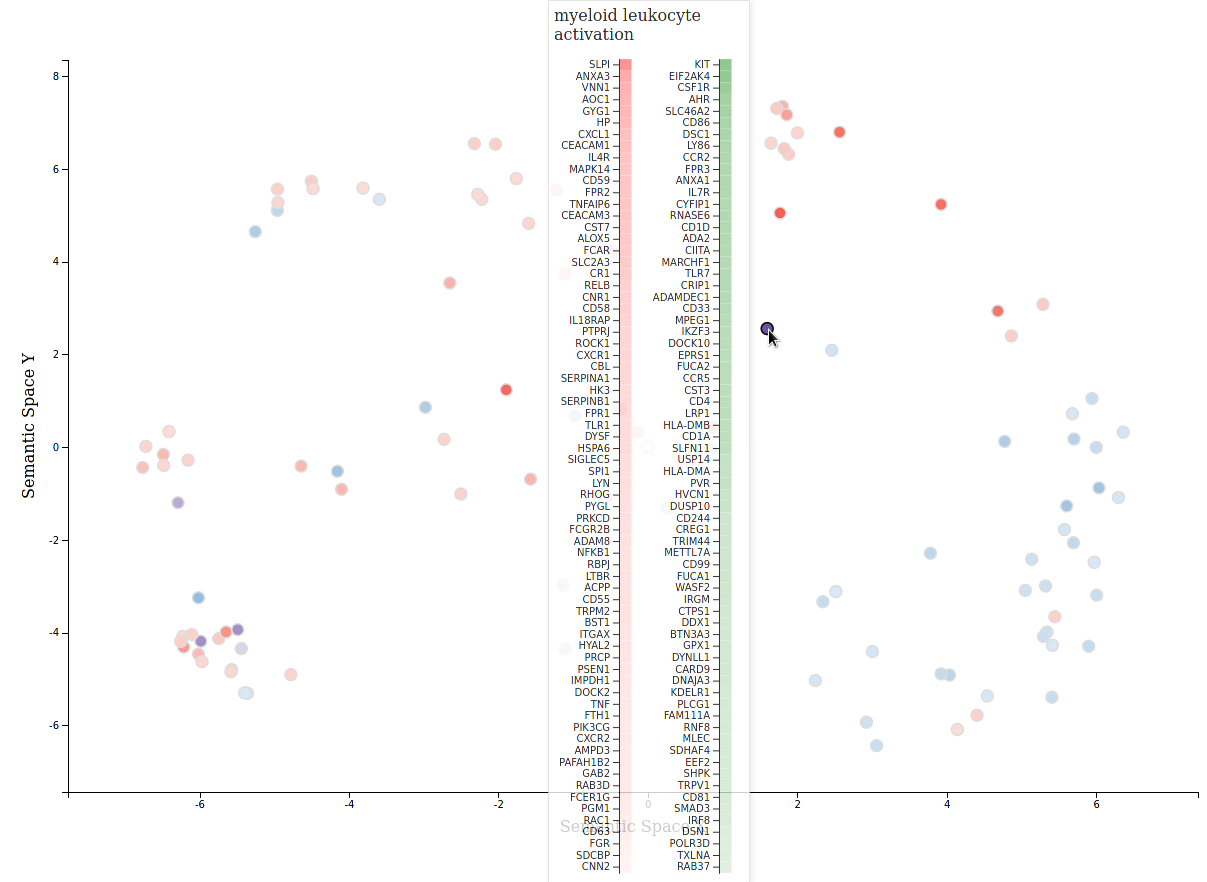
revigo_forcegraph(data_dir)revigo_scatterplot also supports two analysis results:
revigo_scatterplot(data_dir)