CpG_Me is a WGBS pipeline that takes you from raw fastq files to CpG methylation count matrices (Bismark cytosine reports), where it preprocesses data to remove biases and provides ample QC/QA. Scripts are available for both paired end (PE) and single end (SE) sequencing approaches. The extracted CpG methylation count matrices can be then be used for the identification of differentially methylated regions (DMRs) through the accompanying DMRichR workflow.
- Overview
- Quick Start
- Installation
- Chastity Filtering
- Merging Lanes
- Correcting for Methylation Bias (m-bias)
- Paired End (PE) Sequencing
- Single End (SE) Sequencing
- QC Report
- Citation
- Publications
- Acknowledgements
A single command line call performs the following steps for all samples:
- Trim adapters and methylation bias
- Screen for contaminating genomes
- Align
- Remove PCR duplicates
- Calculate nucleotide frequencies (coverage)
- Extract CpG methylation
- Merge symmetric CpG sites
- Individual sample quality control and analysis (QC/QA)
- Legacy file (DSS/DMRfinder) conversion
A final command line call generates html QC/QA reports for all samples that easily enables the identification of failed samples and specifically what went wrong.
Assuming everything is installed and that you have paired-end 150 bp reads from the NovaSeq, where Swift's Accel-NGS Methyl-Seq Kit kit was used for library preparation, all you need to do is change to your working directory and run the following command for the human genome:
sbatch --array=1-96 /share/lasallelab/programs/CpG_Me/Paired-end/CpG_Me_PE_controller.sh hg38
Once your alignments are complete, you can generate the overall QC/QA report with the following command:
sbatch /share/lasallelab/programs/CpG_Me/Paired-end/CpG_Me_PE_QC.sh
This workflow utilizes the following packages, which need to be installed and in your path:
I recommend using Bioconda to install and manage the package updates, which can be accomplished by:
conda install -c bioconda trim-galore bismark bowtie2 samtools fastq-screen multiqc
Bisulfite converted genomes will also have to be created and placed in an external folder for the genome of interest as well as the genomes you would like to use to screen for contamination. This can be accomplished by using bismark_genome_preperation, which is detailed in the Bismark docs, and example scripts are available in the Genome_preperation folder of this repository. These scripts expect that each bisulfite converted genome is located in a genomes folder, which contains a folder for each genome within it (i.e. hg38). However, you can also download the prepared indices for a number of genomes via FastQ Screen with the command fastq_screen --bisulfite --get_genomes.
The genome folder structure should appear as:
├── genomes
│ ├── hg38
│ │ ├── Bisulfite_Genome
│ │ │ ├── CT_conversion
│ │ │ ├── GA_conversion
│ │ ├── Bowtie2
│ │ ├── hg38.fa
│ │ ├── hg38.fa.fai
│ ├── mm10
│ │ ├── Bisulfite_Genome
│ │ │ ├── CT_conversion
│ │ │ ├── GA_conversion
│ │ ├── Bowtie2
│ │ ├── mm10.fa
│ │ ├── mm10.fa.fai
The paths will also need to be changed in the controller, switch, and QC scripts through the mainPath variable, where for our environment they begin with /share/lasallelab/ and the scripts themselves are located in the programs folder. Finally, the paths will also need to be changed in fastq_screen.conf and multiqc_config.yaml configuration files.
The overall folder structure should appear as:
├── programs
│ ├── CpG_Me
│ │ ├── Genome-preperation
│ │ ├── Paired-end
│ │ ├── Single-end
│ │ ├── fastq_screen.conf
│ │ ├── Bismark_to_Permeth_DSS.py
│ │ ├── README.md
│ │ ├── LICENSE
├── genomes
│ ├── hg38
│ ├── mm10
Finally, if you are interested in using the output with WGBS_tools or DMRfinder, the Bismark_to_Permeth_DSS.py script functions as a file converter. If you do not wish to use this file converter, then the final calls in both the switch and controller scripts should be deleted and you should also remove the jid6=$ and | cut -d " " -f 4) part of the QC/QA call in the controller script.
This workflow assumes your data is Illumina quality/chastity filtered, which service providers these days will do by default, so this step is a vestige for older HiSeq data, and is something you probably don't need to worry about for new datasets.
You can check by using the following command, where file.fastq.gz represents your file:
zcat BL001.fastq.gz | head -n 50
Essentially, you want to make sure all your reads contain :N: and none contain :Y:. If your reads aren’t chastity filtered you can accomplish this on command line via the command below, where you change BL001 to your sample name (see ref):
zcat BL001*fastq.gz | zgrep -A 3 '^@.* [^:]*:N:[^:]*:' | zgrep -v "^--$" | gzip > BL001_filtered.fq.gz
For large-scale studies, there are often more samples than can fit on a single lane of sequencing. Even the NovaSeq has its limits and we generally recommend not to pool more than 48 samples per a NovaSeq lane for this low-coverage WGBS workflow. However, lane effects are a significant source of batch effects in any high-throughput sequencing experiment. In order to combat this source of technical bias, which can produce an effect stronger than biological signal, we create a large library pool (up to 96 samples with dual indices) and then repeatedly sequence that library pool across multiple lanes. A MiSeq run of this pool is also a great QC/QA step that helps with balancing the library pool before the large sequencing commitment.
Once you have your sequencing results, the most straightforward approach to merging the results of multiple lanes of data for the same sample is as follows (see ref):
- Check for the right number of unique sample IDs for both R1 and R2
ls -1 *R1*.gz | awk -F '_' '{print $1}' | sort | uniq | wc -l
ls -1 *R2*.gz | awk -F '_' '{print $1}' | sort | uniq | wc -l
- Create file of unique IDs based on _ delimiter and first string (use file for CpG_Me)
ls -1 *fastq.gz | awk -F '_' '{print $1}' | sort | uniq > task_samples.txt
- Test merge commands for each read (look over each one carefully)
for i in `cat ./task_samples.txt`; do echo cat $i\_*_R1_001.fastq.gz \> $i\_1.fq.gz; done
for i in `cat ./task_samples.txt`; do echo cat $i\_*_R2_001.fastq.gz \> $i\_2.fq.gz; done
- Use merge commands for each read by removing echo and the escape character on >
for i in `cat ./task_samples.txt`; do cat $i\_*_R1_001.fastq.gz > $i\_1.fq.gz; done
for i in `cat ./task_samples.txt`; do cat $i\_*_R2_001.fastq.gz > $i\_2.fq.gz; done
Now, not only are your samples merged across lanes, but you now also have your task_samples.txt file for the next steps. If your data is single end then you need to modify accordingly, where you will also need to slightly modify the task_samples.txt file after too.
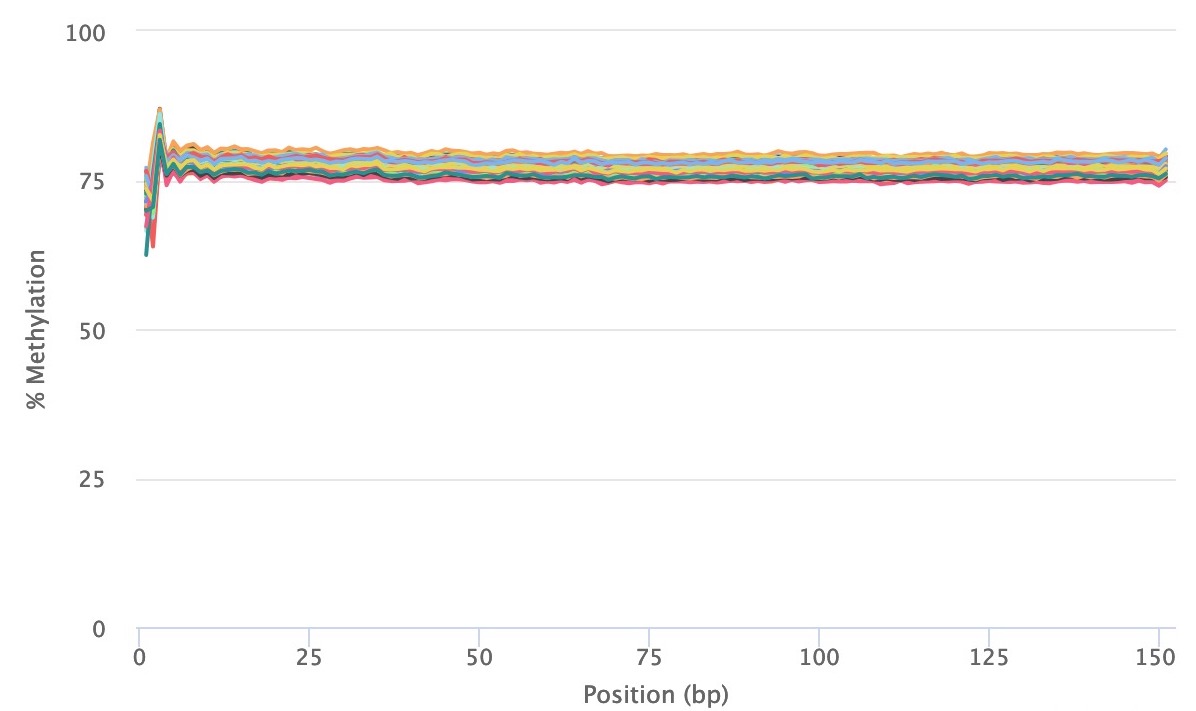
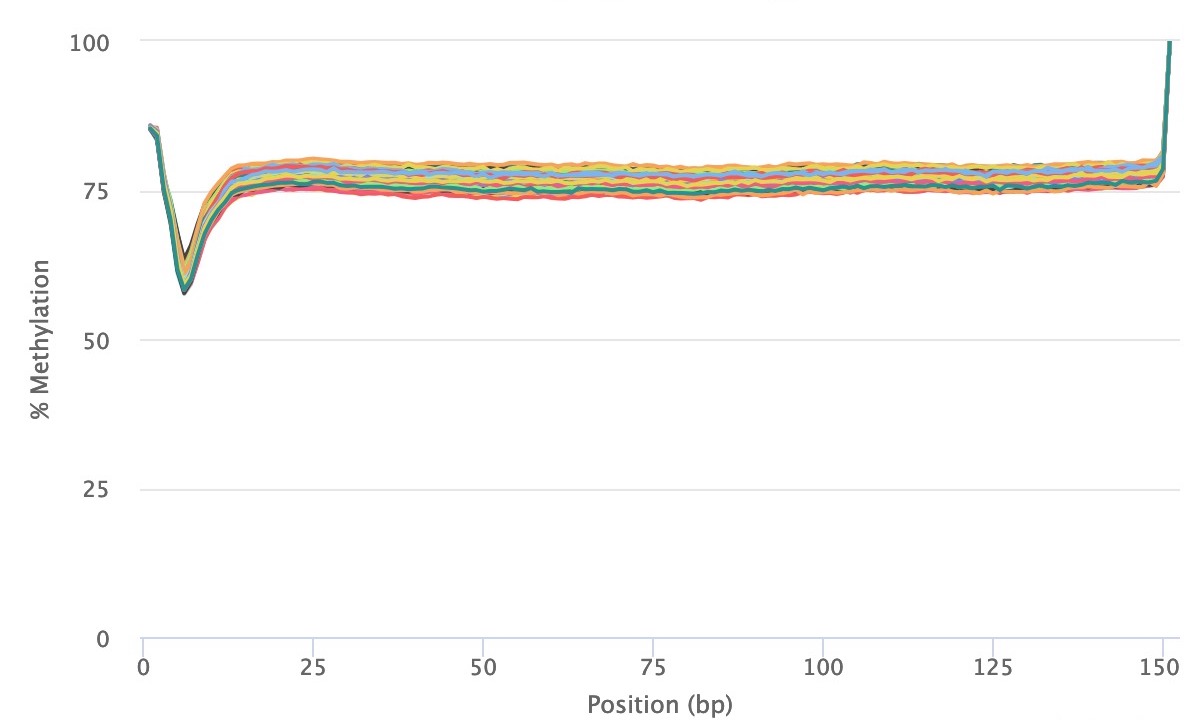
Methylation bias (m-bias) is a technical artifact where the 5' and 3' ends of reads contain artificial methylation levels due to the library preparation method (see Figure 2 in Hansen et al.). One example is the random priming used in post-bisulfite adapter tagging (PBAT) methods (read more here). In paired-end sequencing approaches, the m-bias can also differ between reads 1 and 2 (read more here). Therefore, it is important to always examine for this bias in the MultiQC reports. CpG m-bias can be used to guide trimming options, while CpH m-bias can be used to judge for incomplete bisulfite conversion. In our experience, we have come across the following parameters, although we recommend to examine every dataset, particularly when trying a new library preparation method or sequencing platform. In paired end approaches, the 5' end of read 2 tends to show the largest m-bias.
To address m-bias, the following parameters should be customized in the CpG_Me_switch.sh script:
| Library preparation kit | clip_r1 | clip_r2 | three_prime_clip_r1 | three_prime_clip_r2 |
|---|---|---|---|---|
| Accel-NGS Methyl-Seq Kit (Swift) | 10 | 20 | 10 | 10 |
| TruSeq DNA Methylation Kit (EpiGnome) | 8 | 20 | 8 | 8 |
| Library preparation kit | clip_r1 | three_prime_clip_r1 |
|---|---|---|
| TruSeq DNA Methylation Kit (EpiGnome) | 8 | 8 |
| MethylC-Seq (Original Method) | 7 | 10 |
- Create a parent directory for the project
- Within that parent project directory, add a text file called “task_samples.txt”, where each new line contains the entire sample name exactly as it appears on the fastq read pair files, aside from the end part (“_1.fq.gz” or “_2.fq.gz”). Only name a sample once, NOT twice, and make sure it is .fq.gz and not fastq.gz. Also, if you’re using excel or a windows desktop, you will need to change the linebreaks from windows to unix, which can be done using BBedit (File > Save As... > Line Breaks > Unix) or on command line (but make sure the files have different names):
awk '{ sub("\r$", ""); print }' task_samples_windows.txt > task_samples.txt
- Within that parent directory create a folder called “raw_sequences” that contains all raw paired fastq files (.fq.gz)
Overall, the directory tree structure should be the following:
├── Project
│ ├── raw_sequences
│ │ ├── sample1_1.fq.gz
│ │ ├── sample1_2.fq.gz
│ │ ├── sample2_1.fq.gz
│ │ ├── sample2_2.fq.gz
│ ├── task_samples.txt
- Ensure the trimming options in the switch script are appropriate for the Methylation Bias (m-bias) of your library preparation method as well as your sequencing chemistry. The HiSeq and MiSeq series of sequencers use a 4 color chemistry, while NovaSeq and NextSeq series use a 2 color chemistry. For a 4 color chemistry you should use
--quality 20, while for 2 color chemistry you should use--2colour 20.
Now with that structure in place it’s ready to run, so FROM the parent directory, modify and run this command:
sbatch --array=1-12 /share/lasallelab/programs/CpG_Me/Paired-end/CpG_Me_PE_controller.sh hg38
Let’s break this apart:
- sbatch is how you submit a job to a HPCC with a slurm workload manager
- --array=12 lets you specify the number of samples, as well as subset. Here we are running samples 1 to 12. You could run select samples using the following format --array=2,4-12
- The next call is the location of the executable shell script that will schedule all jobs with proper resources and dependencies on a per sample basis
- Genome (hg38, rheMac8, mm10, rn6)
For single end sequencing, follow the same approach as paired end with minor changes.
The directory should appear as:
├── Project
│ ├── raw_sequences
│ │ ├── sample1.fq.gz
│ │ ├── sample2.fq.gz
│ ├── task_samples.txt
The calls to the scripts would be:
sbatch --array=1-12 /share/lasallelab/programs/CpG_Me/Single-end/CpG_Me_SE_controller.sh hg38
There is also a final html QC report that should be run AFTER all samples have finished, which you also need to launch from the working directory. To generate the QC report for paired end sequencing data, the command is:
sbatch /share/lasallelab/programs/CpG_Me/Paired-end/CpG_Me_PE_QC.sh
To generate the QC report for single end sequencing data, the command is:
sbatch /share/lasallelab/programs/CpG_Me/Single-end/CpG_Me_SE_QC.sh
An example report for single end sequencing is available in the Examples folder. There is currently a minor glitch in the paired end reports, where the temporary files for the different reads create empty columns. This can be fixed by clicking on the configure columns button above the general statistics table and re-selecting one of the visibile columns. Also, these reports can be customized by modifying the multiqc_config.yaml files for the paired end and single end pipelines.
If you use CpG_Me in published research please cite the 2 following articles:
Laufer BI, Hwang H, Vogel Ciernia A, Mordaunt CE, LaSalle JM. Whole genome bisulfite sequencing of Down syndrome brain reveals regional DNA hypermethylation and novel disease insights. Epigenetics, 2019. doi: 10.1080/15592294.2019.1609867
Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics, 2011. doi: 10.1093/bioinformatics/btr167
The following publications utilize CpG_Me and DMRichR:
Wöste M, Leitão E, Laurentino S, Horsthemke B, Rahmann S, Schröder C. wg-blimp: an end-to-end analysis pipeline for whole genome bisulfite sequencing data. bioRxiv preprint. doi: 10.1101/859900
Mordaunt CE, Jianu JM, Laufer BI, Zhu Y, Dunaway KW, Bakulski KM, Feinberg JI, Volk HE, Lyall K, Croen LA, Newschaffer CJ, Ozonoff S, Hertz-Picciotto I, Fallin DM, Schmidt RJ, LaSalle JM. Cord blood DNA methylome in newborns later diagnosed with autism spectrum disorder reflects early dysregulation of neurodevelopmental and X-linked genes. bioRxiv preprint. doi: 10.1101/850529
Lopez SJ, Laufer BI, Beitnere U, Berg E, Silverman JL, Segal DJ, LaSalle JM. Imprinting effects of UBE3A loss on synaptic gene networks and Wnt signaling pathways. Human Molecular Genetics, 2019. doi: 10.1093/hmg/ddz221
Vogel Ciernia A*, Laufer BI*, Hwang H, Dunaway KW, Mordaunt CE, Coulson RL, Yasui DH, LaSalle JM. Epigenomic convergence of genetic and immune risk factors in autism brain. Cerebral Cortex, 2019. doi: 10.1093/cercor/bhz115
Laufer BI, Hwang H, Vogel Ciernia A, Mordaunt CE, LaSalle JM. Whole genome bisulfite sequencing of Down syndrome brain reveals regional DNA hypermethylation and novel disease insights. Epigenetics, 2019. doi: 10.1080/15592294.2019.1609867
The author would like to thank Matt Settles from the UC Davis Bioinformatics Core for examples of tidy code and his suggestion of using a case statement to optimize the resource use of the different parts of this workflow on a high-performance computing cluster. Finally, I would like to thank Ian Korf for invaluable discussions related to the bioinformatic approaches utilized in this repository.