Enrich Your Differentially Methylated Region (DMR) Analysis with the Tidyverse
Website: ben-laufer.github.io/DMRichR/
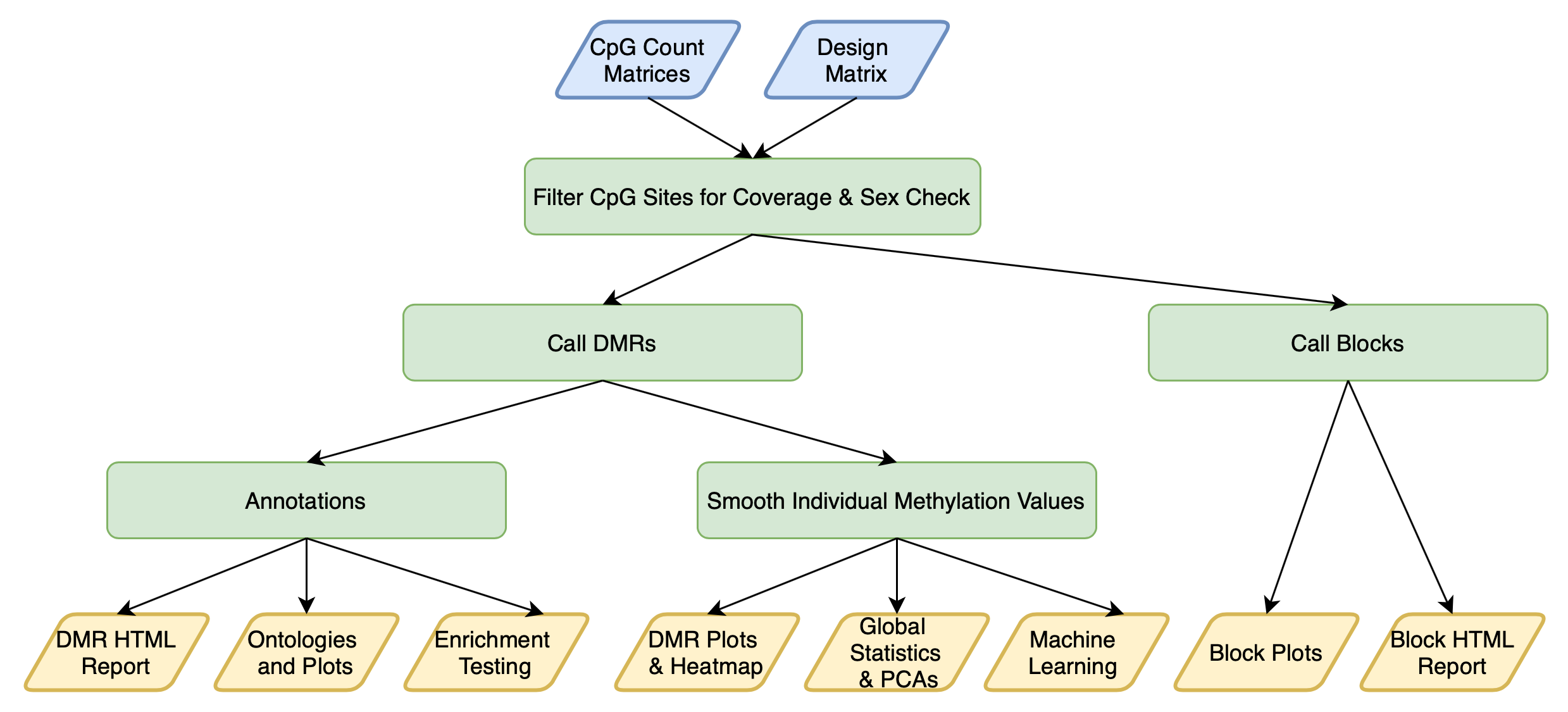
DMRichR is an R package and executable for the preprocessing, statistical analysis, and visualization of differentially methylated regions (DMRs) and global methylation levels from CpG count matrices (Bismark cytosine reports). These files can be generated from your own pipeline or through the CpG_Me pipeline.
DMRichR enables the analysis of data from whole genome bisulfite sequencing (WGBS), enzymatic methyl-seq (EM-seq), and reduced representation bisulfite sequencing (RRBS). The overarching theme of DMRichR is the synthesis of popular Bioconductor R packages for the analysis of genomic data with the tidyverse philosophy of R programming.
DMRichR::DM.R() is a single function that performs all of the following steps:
You can install the package using the following code:
if(!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
if(!requireNamespace("remotes", quietly = TRUE))
install.packages("remotes")
Sys.setenv("R_REMOTES_NO_ERRORS_FROM_WARNINGS" = TRUE)
BiocManager::install("ben-laufer/DMRichR")
If you are running into installation errors with the package dependencies, make sure that you have bioconductor v3.16 and R v4.2 installed. macOS users will have to install XQuartz manually or through Homebrew using brew install xquartz --cask.