Assembler benchmark for ONT MinION data
Authors: Carlos de Lannoy, data from Loman Labs
Generated using poreTally, a benchmarking tool. For an interactive version of this report, download REPORT.html from this repository.
Abstract
The MinION is a portable DNA sequencer that generates long error-prone reads. As both the hardware and analysis software are updated regularly, the most suitable pipeline for subsequent analyses of a dataset generated with a given combination of hardware and software for a given organism is not always clear. Here we present a benchmark for a selection of de novo assemblers available to MinION users, on a read set of Escherichia coli. This benchmark is based on a benchmarking routine, designed to facilitate easy replication on a read set of choice and addition of other de novo assembly pipelines.Methods
Readset quality assessment
Reads in this dataset were generated on a Minion with FLO-MIN106 flowcell with SQK-RAD002 kit. The reads were basecalled using Albacore 0.8.4. Prior to assembly, the quality of the untreated readset was analysed using NanoPlot (version: 1.13.0) and mapped using Minimap2 (version: 2.10-r764-dirty).Assembly pipelines
canu
Canu is a complete OLC assembly pipeline that was shown to work well for the assembly of error-prone reads. It performs a pre-assembly read correction, read trimming, assembly using the minhash alignment process (MHAP) and ultimately a consensus finding step.
Included tools:
- canu (version: snapshot v1.7 +0 changes (r8692 c9ef9219a265e0bbe3a311cca7d28aa02b7517d3))
Used command:
${CANU} -d ${INT}/assembler_results/canu -p canu_assembly maxThreads=${NB_THREADS} useGrid=false genomeSize=$REFGENOME_SIZE -nanopore-raw ${INT}/all_reads.fasta
cp ${INT}/assembler_results/canu/canu_assembly.contigs.fasta ${INT}/assembler_results/all_assemblies/canu.fasta
smartdenovo
SMARTdenovo is a long read OLC assembly pipeline that was originally intended to work with PacBio reads, but has been shown to produce assemblies of reasonably high continuity from MinION reads as well.
Included tools:
- SMARTdenovo (version: none defined)
Used command:
${SMARTDENOVO} -p ${INT}/assembler_results/smartdenovo/smartdenovo_assembly ${INT}/all_reads.fasta > ${INT}/assembler_results/smartdenovo/smartdenovo_assembly.mak && (make -f ${INT}/assembler_results/smartdenovo/smartdenovo_assembly.mak)
if [ -e ${INT}/assembler_results/smartdenovo/smartdenovo_assembly.cns ]; then
cp ${INT}/assembler_results/smartdenovo/smartdenovo_assembly.cns ${INT}/assembler_results/all_assemblies/smartdenovo.fasta
elif [ -e ${INT}/assembler_results/smartdenovo/smartdenovo_assembly.dmo.lay.utg ]; then
cp ${INT}/assembler_results/smartdenovo/smartdenovo_assembly.dmo.lay.utg ${INT}/assembler_results/all_assemblies/smartdenovo.fasta
fiminimap2 miniasm nanopolish
Minimap2 is a fast all-vs-all mapper of reads that relies on sketches of sequences, composed of minimizers. Miniasm uses the found overlaps to construct an assembly graph. As a consensus step is lacking in this pipeline, post-assembly polishing is often required. In this case, Nanopolish was used.
Included tools:
- minimap2 (version: <${MINIMAP2} -V>)
- miniasm (version: <${MINIASM} -V>)
- nanopolish (version: <${NANOPOLISH} --version | grep -Po '(?<=nanopolish version ).+'>)
Used command:
${MINIMAP2} -x ava-ont -t ${NB_THREADS} ${INT}/all_reads.fastq ${INT}/all_reads.fastq | gzip -1 > ${INT}/assembler_results/minimap2_miniasm_nanopolish/minimap2.paf.gz && (${MINIASM} -f ${INT}/all_reads.fastq ${INT}/assembler_results/minimap2_miniasm_nanopolish/minimap2.paf.gz > ${INT}/assembler_results/minimap2_miniasm_nanopolish/minimap2_miniasm.gfa) awk '/^S/{print ">"$2"\n"$3}' ${INT}/assembler_results/minimap2_miniasm_nanopolish/minimap2_miniasm.gfa | fold > ${INT}/assembler_results/minimap2_miniasm_nanopolish/minimap2_miniasm.fasta${TOOL_DIR}/scripts/other/nanopolish_std.sh ${INT}/assembler_results/minimap2_miniasm_nanopolish/minimap2_miniasm.fasta ${INT}/extended_parameters.config ${INT}/all_reads.fastq
cp ${INT}/assembler_results/minimap2_miniasm_nanopolish/nanopolish/minimap2_miniasm_nanopolish.fasta ${INT}/assembler_results/all_assemblies/minimap2_miniasm_nanopolish.fasta
flye
Flye uses A-Bruijn graphs to assemble long error-prone reads. To do so, it follows arbitrary paths through the assembly graph and constructs new assembly graphs from these paths.
Included tools:
- flye (version: 2.3.3-g47cdd0b)
Used command:
$FLYE --nano-raw ${INT}/all_reads.fastq --genome-size ${REFGENOME_SIZE} --out-dir ${INT}/assembler_results/flye/ --threads ${NB_THREADS}
cp ${INT}/assembler_results/flye/scaffolds.fasta ${INT}/assembler_results/all_assemblies/flye.fasta
minimap2 miniasm
Minimap2 is a fast all-vs-all mapper of reads that relies on sketches of sequences, composed of minimizers. Miniasm uses the found overlaps to construct an assembly graph. As a consensus step is lacking in this pipeline, post-assembly polishing is often required.
Included tools:
- minimap2 (version: 2.10-r764-dirty)
- miniasm (version: 0.2-r168-dirty)
Used command:
${MINIMAP2} -x ava-ont -t ${NB_THREADS} ${INT}/all_reads.fastq ${INT}/all_reads.fastq | gzip -1 > ${INT}/assembler_results/minimap2_miniasm/minimap2.paf.gz && (${MINIASM} -f ${INT}/all_reads.fastq ${INT}/assembler_results/minimap2_miniasm/minimap2.paf.gz > ${INT}/assembler_results/minimap2_miniasm/minimap2_miniasm.gfa) awk '/^S/{print ">"$2"\n"$3}' ${INT}/assembler_results/minimap2_miniasm/minimap2_miniasm.gfa | fold > ${INT}/assembler_results/minimap2_miniasm/minimap2_miniasm.fasta
cp ${INT}/assembler_results/minimap2_miniasm/minimap2_miniasm.fasta ${INT}/assembler_results/all_assemblies/minimap2_miniasm.fasta
Assembly quality assessment
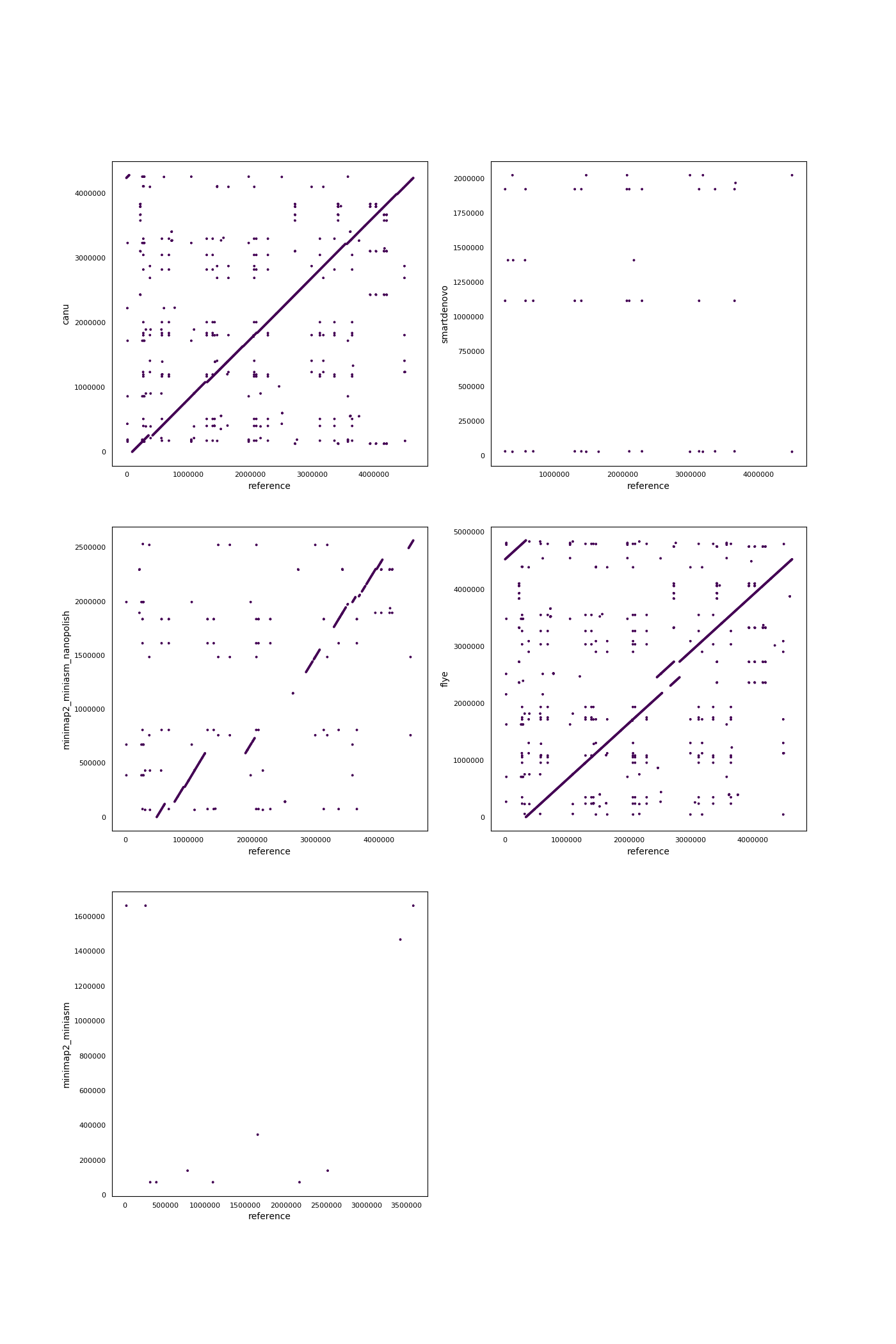
Produced assemblies were analyzed and compared on continuity and agreement with the reference genome. Quast (version: 4.6.2) was used to determine a wide array of quality metrics in both quality categories and produce synteny plots. To elucidate any bias in the occurence of certain sequences, 5-mers in the assemblies and the reference genomes were compared using Jellyfish (version: 2.2.9). Finally, results were summarized using MultiQC.Results
General Statistics
| Total length | N50 | indels per 100 kbp | CPU time | mismatches per 100 kbp | Genome fraction | |
|---|---|---|---|---|---|---|
| smartdenovo | 2.95478e+06 | 457841 | 3776.75 | 0:27:40 | 3692.82 | 0.154 |
| minimap2_miniasm_nanopolish | 2.56604e+06 | 314671 | 3255.26 | 1 day, 4:26:31 | 2893.55 | 32.717 |
| minimap2_miniasm | 2.50698e+06 | 307611 | 3881.88 | 0:00:12 | 2886.53 | 0.065 |
| canu | 4.28217e+06 | 824286 | 2473.37 | 1 day, 16:39:33 | 701.92 | 95.605 |
| flye | 4.85564e+06 | 1.35387e+06 | 1717.34 | 0:46:41 | 920.91 | 99.563 |
Readset quality
| Value | N | % | ||
|---|---|---|---|---|
| Mean read quality | 10.8 | mismatches | 872 | 10.9 |
| Median read quality | 11.2 | deletions | 910 | 11.38 |
| Median read length | 20763 | insertions | 258 | 3.23 |
| Mean read length | 34946.1 | matches | 5958 | 74.49 |
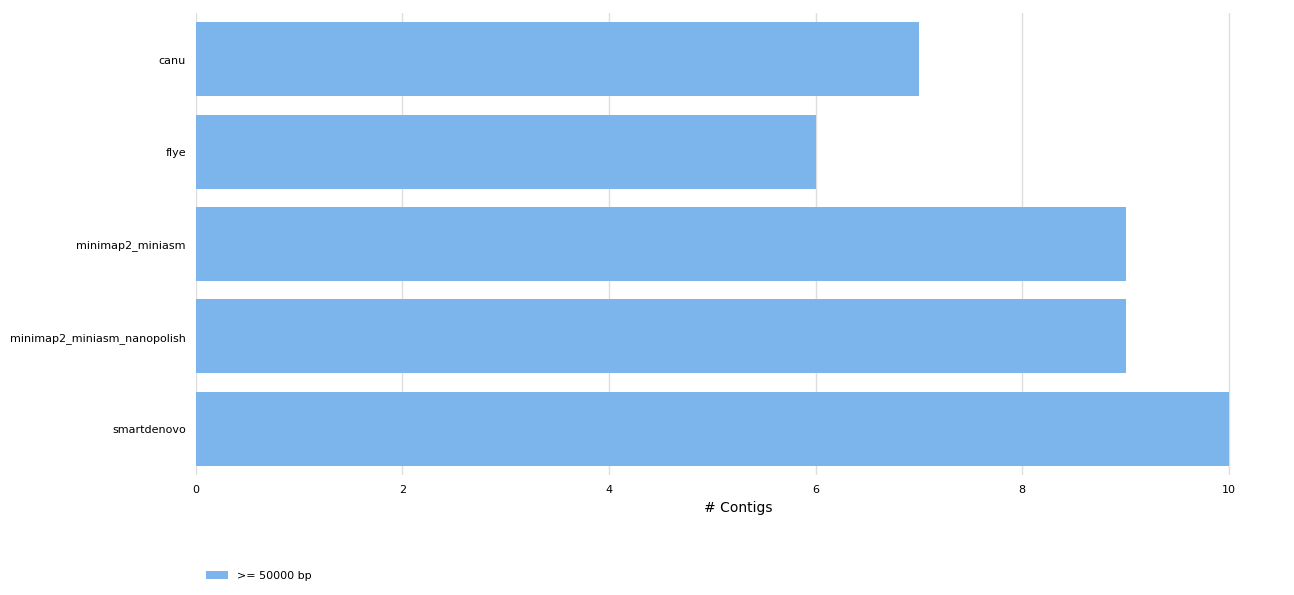
QUAST
Assembly Statistics
| Length (Mbp) | N75 (Kbp) | Largest contig (Kbp) | L50 (K) | N50 (Kbp) | L75 (K) | Genome Fraction | Indels /100Kbp | Mismatches /100Kbp | Genes | Genes (partial) | Misas- semblies | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| smartdenovo | 2.95478e+06 | 204859 | 1.19863e+06 | 2 | 457841 | 5 | 0.154 | 3776.75 | 3692.82 | 7 | 1 | 0 |
| minimap2_miniasm_nanopolish | 2.56604e+06 | 194314 | 1.18852e+06 | 2 | 314671 | 4 | 32.717 | 3255.26 | 2893.55 | 1427 | 47 | 4 |
| minimap2_miniasm | 2.50698e+06 | 190149 | 1.16168e+06 | 2 | 307611 | 4 | 0.065 | 3881.88 | 2886.53 | 5 | 1 | 0 |
| canu | 4.28217e+06 | 553925 | 1.37499e+06 | 2 | 824286 | 4 | 95.605 | 2473.37 | 701.92 | 4118 | 33 | 0 |
| flye | 4.85564e+06 | 935133 | 1.75901e+06 | 2 | 1.35387e+06 | 3 | 99.563 | 1717.34 | 920.91 | 4287 | 14 | 1 |