This is the implementation (in PyTorch) of the following paper
Harnessing Uncertainty in Domain Adaptation for MRI Prostate Lesion Segmentation
Eleni Chiou, Francesco Giganti, Shonit Punwani, Iasonas Kokkinos, Eleftheria Panagiotaki
International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), 2020.
If you find this code useful for your research, please cite our paper:
@inproceedings{chiou20DA,
title={Harnessing Uncertainty in Domain Adaptation for MRI Prostate Lesion Segmentation},
author={Chiou, Eleni and Giganti, Francesco and Punwani, Shonit and Kokkinos, Iasonas and Panagiotaki, Eleftheria},
booktitle={MICCAI},
year={2020}
}
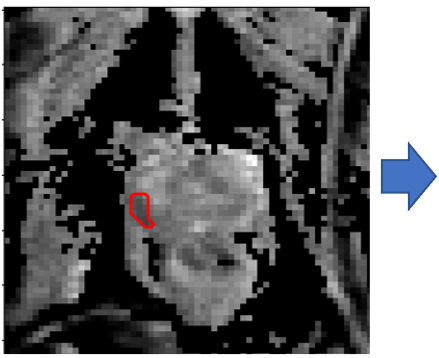
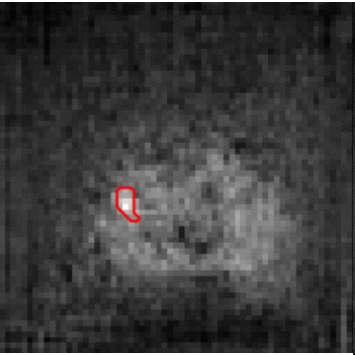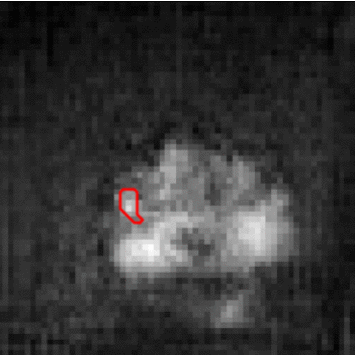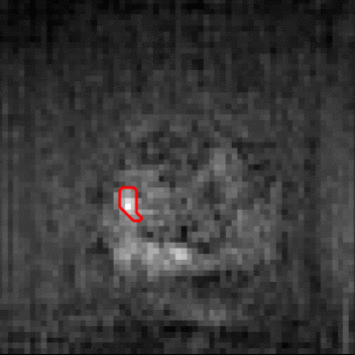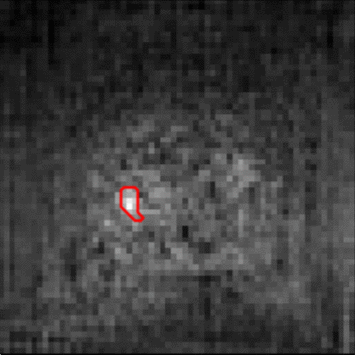
The need for training data can impede the adoption of novel imaging modalities for learning-based medical image analysis. Domain adaptation methods partially mitigate this problem by translating training data from a related source domain to a novel target domain, but typically assume that a one-to-one translation is possible. Our work addresses the challenge of adapting to a more informative target domain where multiple target samples can emerge from a single source sample. In particular we consider translating from mp-MRI to VERDICT, a richer MRI modality involving an optimized acquisition protocol for cancer characterization. We explicitly account for the inherent uncertainty of this mapping and exploit it to generate multiple outputs conditioned on a single input. Our results show that this allows us to extract systematically better image representations for the target domain, when used in tandem with both simple, CycleGAN-based baselines, as well as more powerful approaches that integrate discriminative segmentation losses and/or residual adapters. When compared to its deterministic counterparts, our approach yields substantial improvements across a broad range of dataset sizes, increasingly strong baselines, and evaluation measures.
This code was tested with Python 3.6, PyTorch 0.4.1, and CUDA 8.0/9.0.
-
Install PyTorch.
conda install pytorch=0.4.1 torchvision cuda90 -c pytorch -
Install additional dependencies.
pip install -r requirements.txt -
Clone the repo.
git clone https://github.com/elchiou/DA.git
-
Download the dataset you want to use. The dataset directory should have the following structure.
<data_root>/datasets/trainA/data % images <data_root>/datasets/trainA/seg_masks/ % segmentation masks <data_root>/datasets/trainB/data % images <data_root>/datasets/trainB/seg_masks/ % segmentation masks ... -
Setup the configs/config.yaml file.
-
Train the model.
./scripts/train.sh -
Intermediate image outputs and models are stored in results.
MUNIT implementation is borrowed from here.