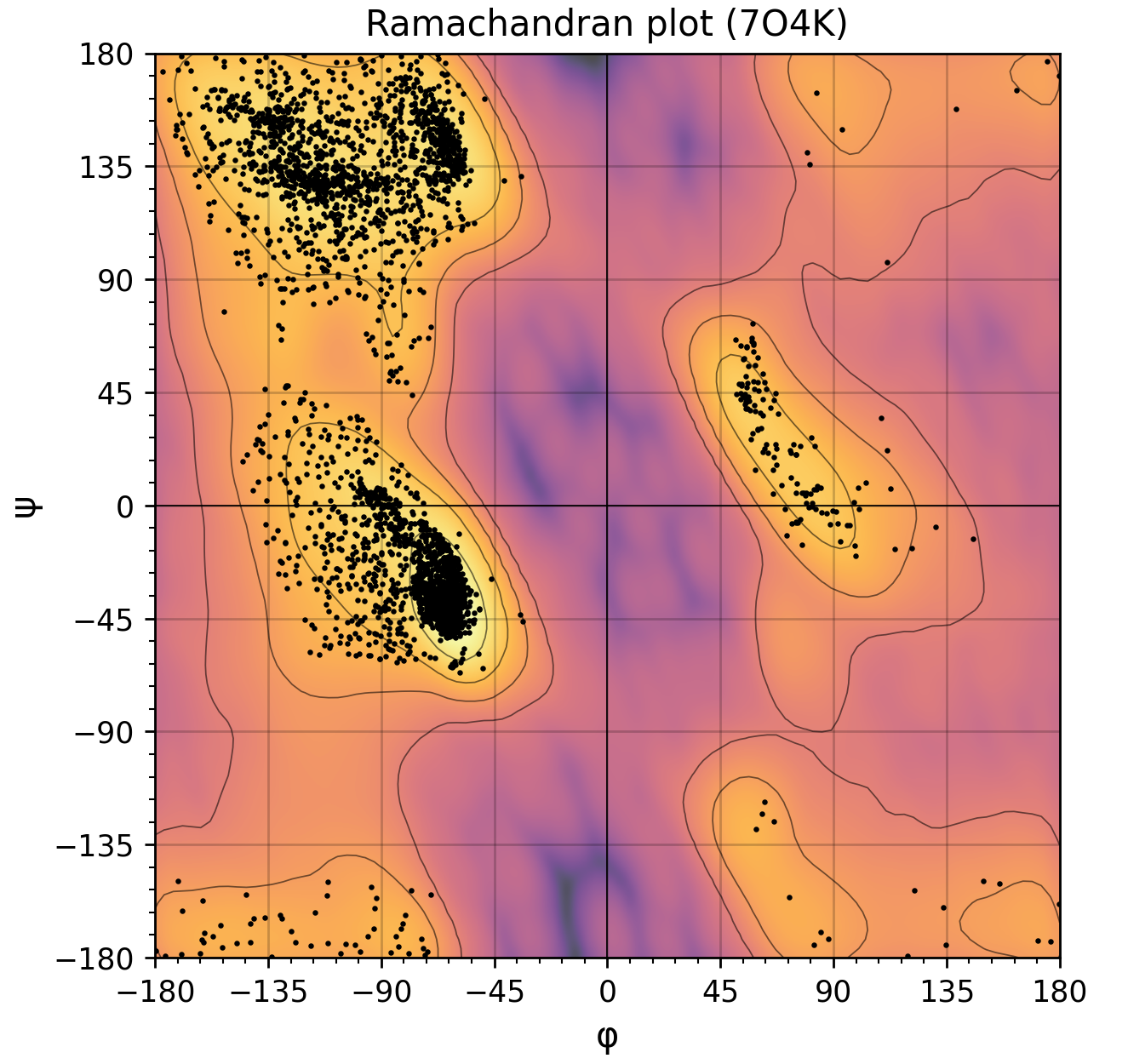
A simple script to generate a Ramachandran plot from structures within pymol.
Over is an example of the generated ramachandran plot using the Yeast TFIIH structure from PDB.
- Download the repository to your computer. Store the code in $pwd (preferred). Find this directory by running,
PyMOL> pwd
The repository can be downloaded either using Git or Download ZIP directly in Github. Using git this can be done using
$ git clone https://github.com/exTerEX/pymol-ramachandran.git /path/to/pwd
- Run script in pymol (example assume script is in $pwd)
$ run ramachandran.py
- Generatet a Ramachandran plot. Fetch one or more structures in pymol and run
$ ramaplot()
or specify which model/residues to be used to generate the plot
$ ramaplot(model="(pdb-id)", resn="(residue)")
or use multiple models/residues
$ ramaplot(model=["(pdb-id)", "(pdb-id)", ...], resn=["(residue)", "(residue)", ...])
This project is licensed under ECL-2.0. See LICENSE for more information.