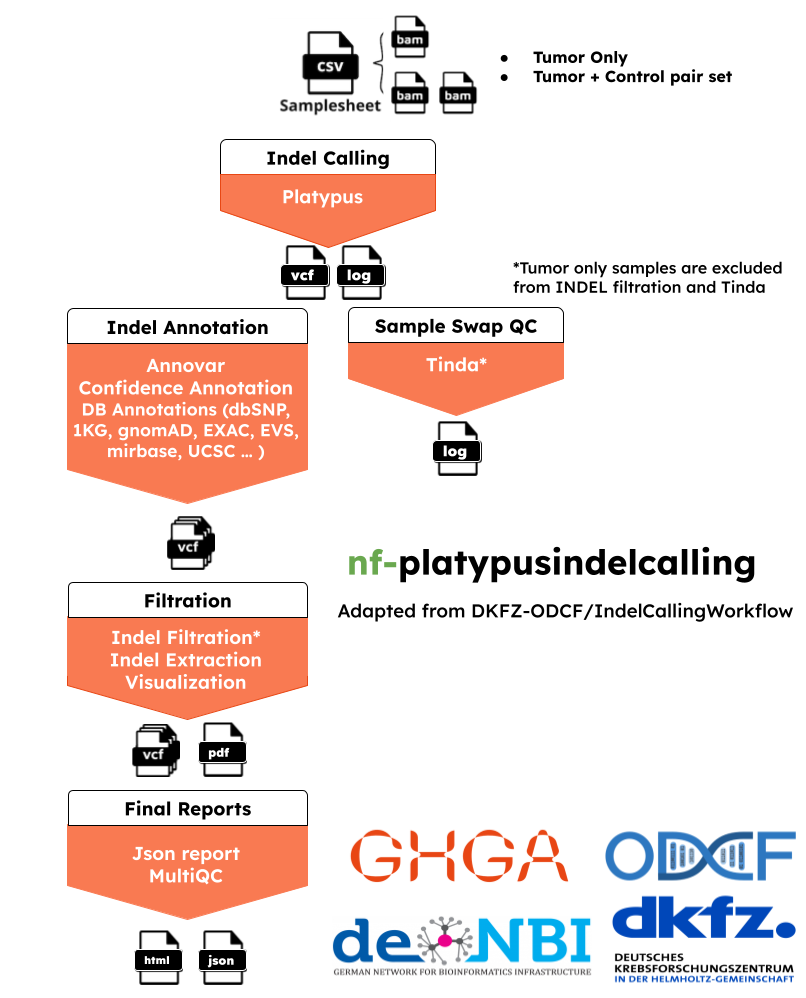
nf-platypusindelcalling:A Platypus-based insertion/deletion-detection workflow with extensive quality control additions. The workflow is based on DKFZ - ODCF OTP Indel Calling Pipeline.
For now, this workflow is only optimal to work in ODCF Cluster. The config file (conf/dkfz_cluster.config) can be used as an example. Running Annotation, DeepAnnotation, Filter and Tinda steps are optional and can be turned off using [runIndelAnnotation, runIndelDeepAnnotation, runIndelVCFFilter, runTinda] parameters sequential.
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It uses Docker/Singularity containers making installation trivial and results highly reproducible. The Nextflow DSL2 implementation of this pipeline uses one container per process which makes it much easier to maintain and update software dependencies.
This nextflow pipeline is the transition of DKFZ-ODCF/IndelCallingWorkflow.
Important Notice: The whole workflow is only ready for DKFZ cluster users for now, It is strongly recommended to them to read whole documentation before usage. This workflow works better with nextflow/22.07.1-edge in the cluster, It is recommended to use >22.07.1.
The pipeline has 6 main steps: Indel calling using platypus, basic annotations, deep annotations, filtering, sample swap check and multiqc report.
-
Indel Calling:
Platypus (
Platypus) : Platypus tool is used to call variants using local realignmnets and local assemblies. It can detect SNPs, MNPs, short indels, replacements, deletions up to several kb. It can be both used with WGS and WES. The tool has been thoroughly tested on data mapped with Stampy and BWA. -
Basic Annotations (--runIndelAnnotation True):
In-house scripts to annotate with several databases like gnomAD, dbSNP, and ExAC.
ANNOVAR (
Annovar) : annotate_variation.pl is used to annotate variants. The tool makes classifications for intergenic, intogenic, nonsynoymous SNP, frameshift deletion or large-scale duplication regions.ENSEMBL VEP('ENSEBL VEP') :can also be used alternative to annovar. Gene annotations will be extracted.
Reliability and confidation annotations: It is an optional ste for mapability, hiseq, selfchain and repeat regions checks for reliability and confidence of those scores.
-
Deep Annotation (--runIndelDeepAnnotation True):
If basic annotations are applied, an extra optional step for number of extra indel annotations like enhancer, cosmic, mirBASE, encode databases can be applied too.
-
Filtering and Visualization (--runIndelVCFFilter True):
It is an optional step. Filtering is only required for the tumor samples with no-control and filtering can only be applied if basic annotation is performed.
Indel Extraction and Visualizations: INDELs can be extracted by certain minimum confidence level
Visualization and json reports: Extracted INDELs are visualized and analytics of INDEL categories are reported as JSON.
-
Check Sample Swap (--runTinda True):
Canopy Based Clustering and Bias Filter, thi step can only be applied into the tumor samples with control.
-
MultiQC (--skipmultiqc False):
Produces pipeline level analytics and reports.
-
Install
Nextflow(>=21.10.3) -
Install any of
DockerorSingularity(you can follow this tutorial) -
Download Annovar and set-up suitable annotation table directory to perform annotation. Example:
annotate_variation.pl -downdb wgEncodeGencodeBasicV19 humandb/ -build hg19Gene annotation is also possible with ENSEMBL VEP tool, for test purposes only, it can be used online. But for big analysis, it is recommended to either download cache file or use --download_cache flag in parameters.
Follow the documentation here
Example:
Download cache
cd $HOME/.vep
curl -O https://ftp.ensembl.org/pub/release-110/variation/indexed_vep_cache/homo_sapiens_vep_110_GRCh38.tar.gz
tar xzf homo_sapiens_vep_110_GRCh38.tar.gz-
Download the pipeline and test it on a minimal dataset with a single command:
git clone https://github.com/ghga-de/nf-platypusindelcalling.git
before run do this to bin directory, make it runnable!:
chmod +x bin/*nextflow run main.nf -profile test,YOURPROFILE --outdir <OUTDIR> --input <SAMPLESHEET>Note that some form of configuration will be needed so that Nextflow knows how to fetch the required software. This is usually done in the form of a config profile (YOURPROFILE in the example command above). You can chain multiple config profiles in a comma-separated string.
- The pipeline comes with config profiles called
dockerandsingularitywhich instruct the pipeline to use the named tool for software management. For example,-profile test,docker.- Please check nf-core/configs to see if a custom config file to run nf-core pipelines already exists for your Institute. If so, you can simply use
-profile <institute>in your command. This will enable eitherdockerorsingularityand set the appropriate execution settings for your local compute environment.- If you are using
singularity, please use thenf-core downloadcommand to download images first, before running the pipeline. Setting theNXF_SINGULARITY_CACHEDIRorsingularity.cacheDirNextflow options enables you to store and re-use the images from a central location for future pipeline runs.
-
Simple test run
nextflow run main.nf --outdir results -profile singularity,dkfz_cluster_38 -
Start running your own analysis!
nextflow run main.nf --input samplesheet.csv --outdir <OUTDIR> -profile <docker/singularity> --config test/institute.config
sample: The sample name will be tagged to the job
tumor: The path to the tumor file
tumor_index: The path to the tumor index file
control: The path to the control file, if there is no control will be kept blank.
control_index: The path to the control index file, if there is no control will be kept blank.
Annotations are optional for the user. All VCF and BED files need to be indexed with tabix and should be in the same folder!
The reference set bundle which is used in PCAWG study can be found and downloaded here. (NOTE: only in hg19)
Basic Annotation Files
- dbSNP INDELs (vcf)
- 1000K INDELs (vcf)
- gnomAD Genome Sites for INDELs (vcf)
- gnomAD Exome Sites for INDELs (vcf)
- EVS variants (vcf)
- ExAC variants (vcf)
- Local Control files WGS (vcf)
- Local Control files WES (vcf)
SNV Reliability Files
- UCSC Repeat Masker region (bed)
- UCSC Mappability regions (bed)
- UCSC Simple tandem repeat regions (bed)
- UCSC DAC Black List regions (bed)
- UCSC DUKE Excluded List regions (bed)
- UCSC Hiseq Deep sequencing regions (bed)
- UCSC Self Chain regions (bed)
Deep Annotation Files
- UCSC Enhangers (bed)
- UCSC CpG islands (bed)
- UCSC TFBS noncoding sites (bed)
- UCSC Encode DNAse cluster (bed.gz)
- snoRNAs miRBase (bed)
- miRBase (bed)
- Cosmic coding SNVs (bed)
- miRNA target sites (bed)
- Cgi Mountains (bed)
- UCSC Phast Cons Elements (bed)
- UCSC Encode TFBS (bed)
This pipeline favors the use of igenomes and refgenie. Read the documentaton here to learn more.
For igenomes usage: use genomes GRCh37 (--genome "GRCh37") or GRCh38 (--genome "GRCh38").
For refgenie usage: use genomes GRCh37 (--genome "hg37") or GRCh38 (--genome "hg38").
If not using igenomes or refgenie, --fasta, --fasta_fai, and --chr_prefix need to be spesifed! If --chr_sizes is not provided it will be automatically generated.
The nf-platypusindelcalling pipeline comes with documentation about the pipeline usage and output.
Please read usage document to learn how to perform sample analysis provided with this repository!
nf-platypusindelcalling was originally translated from roddy-based pipeline by Kuebra Narci kuebra.narci@dkfz-heidelberg.de.
The pipeline is originally written in workflow management language Roddy. Inspired github page
The Indel calling workflow was in the pan-cancer analysis of whole genomes (PCAWG) and can be cited in the following publication:
Pan-cancer analysis of whole genomes. The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Nature volume 578, pages 82–93 (2020). DOI 10.1038/s41586-020-1969-6
We thank the following people for their extensive assistance in the development of this pipeline:
- Nagarajan Paramasivam (@NagaComBio) n.paramasivam@dkfz.de
TODO
If you would like to contribute to this pipeline, please see the contributing guidelines.
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.