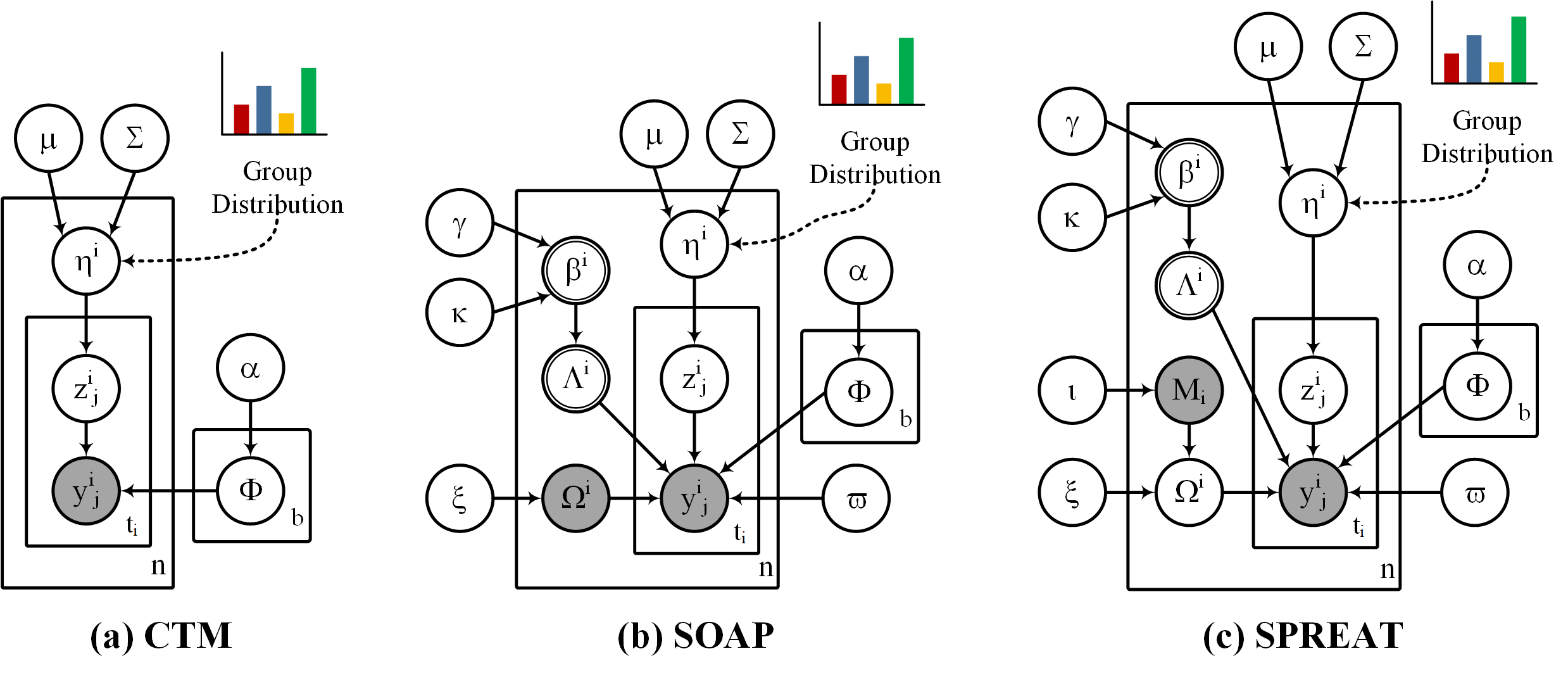
This repo contains an implementation of CHAP (Correlated patHwAy-grouP) package which consists of three hierarchical Bayesian (parametric) mixture models, CTM (Correlated Topic Model), SOAP (Sparse cOrrelated pAthway grouP) and SPREAT (diStributed sParse corRElated pAThway group), that incorporate pathway abundance information to encode each example as a mixture distribution of groups, and each group, in turn, is a mixture of pathways with different mixing proportions. SOAP and SPREAT incorporate supplementary pathway (with different proportions) on top of the pathways provided in pathway datasets to partially resolve noisy pathway data. Moreover, SOAP and SPREAT induce dual sparseness, where we allow an example to select only a few groups and also each group to select its optimum set of pathways. While these models were proposed to model pathways among organims, they can be used for other types of datasets (e.g. natural language processing). We also included LDA (Latent Dirichlet Allocation) which is implemented in scikit-learn.
The codebase is tested to work under Python 3.7. To install the necessary requirements, run the following commands:
pip install -r requirements.txt
Basically, CHAP requires the following distribution and packages:
- Anaconda
- NumPy (== 1.18.5)
- scikit-learn (== 0.23)
- scipy (==1.5.0)
- gensim (==3.8.3)
Please download the following files from Zenodo. Once you have
downloaded the cbt_materials.zip file, unzip it and make sure you obtain the two folders: model/ and dataset/, as
depicted below:
chap_materials/
├── model/
│ ├── ctm.pkl and ctm_[exp_omega | mu | sigma].npz
│ ├── soap.pkl and soap_[exp_phi | mu | sigma].npz
│ ├── spreat.pkl and spreat_[exp_phi | mu | sigma].npz
│ └── ...
└── dataset/
├── biocyc_dictionary.pkl
├── pnas_dictionary.pkl
├── biocyc_features.pkl
├── biocyc_X.pkl, biocyc_text_X.pkl, biocyc_M.pkl
├── golden_X.pkl, golden_text_X.pkl, golden_M.pkl
├── cami_X.pkl, cami_text_X.pkl, cami_M.pkl
├── pnas_X.pkl, pnas_Xtrain.pkl, pnas_Xtest.pkl, pnas_text_X.pkl, pnas_text_Xtrain.pkl, pnas_text_Xtest.pkl
└── ...
A short description of the contents of the above folders is given below.
- "ctm.pkl" and "ctm_[exp_omega | mu | sigma].npz": a pretrained CTM model using biocyc_X data and associated files.
- "soap.pkl" and "soap_[exp_phi | mu | sigma].npz": a pretrained SOAP model using biocyc_X data and associated files.
- "spreat.pkl" and "spreat_[exp_phi | mu | sigma].npz": a pretrained SPREAT model using biocyc_X data and associated files.
- "biocyc_dictionary.pkl": a Dictionary object (from gensim) consists of 2526 distinct tokens (pathways).
- "biocyc_features.pkl": a sample of embeddings (nodes, dimension size). Based on your tests, you need to generate features using pathway2vec.
- "biocyc_X.pkl": BioCyc (v21 tier 2 & 3) dataset of size (9429, 2526).
- "biocyc_text_X.pkl": A list of pathways for BioCyc (v21 tier 2 & 3) dataset of size (9429 samples).
- "biocyc_M.pkl": Supplementary pathways for BioCyc (v21 tier 2 & 3) dataset of size (9429, 2526).
- "golden_X.pkl": Golden dataset of size (63, 2526). First six examples correspond to: AraCyc, EcoCyc, HumanCyc, LeishCyc, TrypanoCyc, and YeastCyc.
- "golden_text_X.pkl": A list of pathways for golden dataset of size (63 samples). First six examples correspond to: AraCyc, EcoCyc, HumanCyc, LeishCyc, TrypanoCyc, and YeastCyc.
- "golden_M.pkl": Supplementary pathways for golden dataset of size (63, 2526). First six examples correspond to: AraCyc, EcoCyc, HumanCyc, LeishCyc, TrypanoCyc, and YeastCyc.
- "cami_X.pkl": CAMI dataset of size (40, 2526).
- "cami_text_X.pkl": A list of pathways for CAMI dataset of size (40 samples).
- "cami_M.pkl": Supplementary pathways for CAMI dataset of size (40, 2526).
- "pnas_dictionary.pkl": a Dictionary object (from gensim) consists of 26561 distinct tokens (words).
- "pnas_X.pkl": PNAS dataset of size (1391, 26561).
- "pnas_Xtrain.pkl": PNAS training dataset of size (1113, 26561).
- "pnas_Xtest.pkl": PNAS test dataset of size (278, 26561).
- "pnas_text_X.pkl": PNAS dataset (a list of strings) of size (1391 samples).
- "pnas_text_Xtrain.pkl": PNAS training dataset (a list of strings) of size (1113 samples).
- "pnas_text_Xtest.pkl": PNAS test dataset (a list of strings) of size (278 samples).
Run the following commands to clone the repository to an appropriate location:
git clone https://github.com/arbasher/cbt
For all experiments, navigate to src folder then run the commands of your choice. For example, to display CHAP's running options use: python main.py --help. It should be self-contained.
For training, we provide few basic examples.
Description about arguments in all examples: --num-components corresponds the number of components (i.e., groups or topics), --top-k indicates the top k features (per component) to be considered for sparseness (SOAP and SPREAT only), --max-sampling is the maximum number of random samplings applied to SOAP and SPREAT only, --vocab-name is the file name for a Dictionary object (from gensim) containing vocabulary tokens (e.g. pathways or words), --ssample-input-size corresponds the size of random subsampled inputs, --fr represents a hyper-parameter (set within the range of (0.5, 1.0]) to indicate the forgotten rate of the previous learned parameters, --delay is another hyper-parameter to down weights early iterations, --batch is batch size, --num-jobs corresponds the number of parallel workers, --max-inner-iter corresponds the number of inner iteration inside a single epoch, --num-epochs corresponds the number of iterations over the training set, and --model-name corresponds the name of the model excluding EXTENSION. The model name will have .pkl extension and an appropriate prefix (e.g. "soap_*.pkl"). The arguments --X-name is the input space of data (e.g. "biocyc_X.pkl" or "pnas_Xtrain.pkl"), --M-name is the supplementary data (same size as X), and --features-name corresponds the features of pathways obtained from pathway2vec.
The results of running all the commands below will be stored in two locations: --mdpath which contains the trained model with additional files and --rspath which has the cost file.
If you wish to train LDA, CTM, SOAP, and SPREAT using a dataset (e.g. "biocyc_X.pkl"), execute the following command:
python main.py --train --soap --spreat --ctm --lda --num-components 200 --top-k 10 --max-sampling 3 --ssample-input-size 0.1 --fr 0.9 --delay 1.0 --X-name "biocyc_X.pkl" --vocab-name "biocyc_dictionary.pkl" --model-name "[model name (without extension)]" --mdpath "[path to the model]" --dspath "[path to the dataset]" --rspath "[path to store the results (e.g. costs)]" --logpath "[path to the log directory]" --batch 50 --max-inner-iter 5 --num-epochs 10 --num-jobs 2
To train SOAP and SPREAT using a dataset (e.g. "biocyc_X.pkl") with collapse2ctm option, execute the following command:
python main.py --train --soap --spreat --collapse2ctm --num-components 200 --max-sampling 3 --ssample-input-size 0.1 --fr 0.9 --delay 1.0 --X-name "biocyc_X.pkl" --vocab-name "biocyc_dictionary.pkl" --model-name "[model name (without extension)]" --mdpath "[path to the model]" --dspath "[path to the dataset]" --rspath "[path to store the results (e.g. costs)]" --logpath "[path to the log directory]" --batch 50 --max-inner-iter 5 --num-epochs 10 --num-jobs 2
To train SOAP and SPREAT using supplementary dataset (e.g. "biocyc_M.pkl") in addition to the original undistorted data (e.g. "biocyc_X.pkl"), enable --use-supplement argument and then execute the following command:
python main.py --train --soap --spreat --use-supplement --top-k 20 --num-components 200 --max-sampling 3 --ssample-input-size 0.1 --fr 0.9 --delay 1.0 --X-name "biocyc_X.pkl" --M-name "biocyc_M.pkl" --vocab-name "biocyc_dictionary.pkl" --model-name "[model name (without extension)]" --mdpath "[path to the model]" --dspath "[path to the dataset]" --rspath "[path to store the results (e.g. costs)]" --logpath "[path to the log directory]" --batch 50 --max-inner-iter 5 --num-epochs 10 --num-jobs 2
Note that the --top-k induces sparseness of pathway distribution over groups.
To train SOAP and SPREAT using features and supplementary dataset (e.g. "biocyc_M.pkl") in addition to the original undistorted data (e.g. "biocyc_X.pkl"), enable --use-supplement and --use-features arguments and then execute the following command:
python main.py --train --soap --spreat --use-supplement --use-features --top-k 20 --num-components 200 --max-sampling 3 --ssample-input-size 0.1 --fr 0.9 --delay 1.0 --X-name "biocyc_X.pkl" --M-name "biocyc_M.pkl" --vocab-name "biocyc_dictionary.pkl" --features-name "biocyc_features.pkl" --model-name "[model name (without extension)]" --mdpath "[path to the model]" --dspath "[path to the dataset]" --rspath "[path to store the results (e.g. costs)]" --logpath "[path to the log directory]" --batch 50 --max-inner-iter 5 --num-epochs 10 --num-jobs 2
Note that the --top-k induces sparseness of pathway distribution over groups.
You can train LDA, CTM, SOAP, and SPREAT using any other datasets (e.g. "pnas_Xtrain.pkl"), execute the following command:
python main.py --train --soap --spreat --ctm --lda --num-components 200 --top-k 10 --max-sampling 3 --ssample-input-size 0.1 --fr 0.9 --delay 1.0 --X-name "pnas_Xtrain.pkl" --vocab-name "pnas_dictionary.pkl" --model-name "[model name (without extension)]" --mdpath "[path to the model]" --dspath "[path to the dataset]" --rspath "[path to store the results (e.g. costs)]" --logpath "[path to the log directory]" --batch 50 --max-inner-iter 5 --num-epochs 10 --num-jobs 2
Also, you can train SOAP and SPREAT using the same dataset with collapse2ctm option:
python main.py --train --soap --spreat --collapse2ctm --num-components 200 --max-sampling 3 --ssample-input-size 0.1 --fr 0.9 --delay 1.0 --X-name "pnas_Xtrain.pkl" --vocab-name "pnas_dictionary.pkl" --model-name "[model name (without extension)]" --mdpath "[path to the model]" --dspath "[path to the dataset]" --rspath "[path to store the results (e.g. costs)]" --logpath "[path to the log directory]" --batch 50 --max-inner-iter 5 --num-epochs 10 --num-jobs 2
For evaluating, we provide few basic examples.
Description about arguments in all examples: --top-k in the evaluation setting entails the top k features (per component) to be considered for for computing the coherence score, --cal-average is a binary value indicating whether to calculate the expected predictive distribution, --vocab-name is the file name for a Dictionary object (from gensim) containing vocabulary tokens (e.g. pathways or words), --batch is batch size, --num-jobs corresponds the number of parallel workers, --num-epochs corresponds the number of iterations over the training set, and --model-name corresponds the name of the learned model excluding PREFIX and EXTENSION. The model name will have .pkl extension. The arguments --X-name is the input space of data (e.g. "cami_X.pkl" or "pnas_Xtest.pkl") and --text-name is the input space of the same data but represented as a list of strings (e.g. "cami_text_X.pkl" or "pnas_text_Xtest.pkl") while the argument --M-name corresponds the supplementary data (same size as X). Finally, --features-name corresponds the features of pathways obtained from pathway2vec.
The results of running all the commands below will be stored in --rspath which contains the score files.
To evaluate pretrained models of LDA, CTM, SOAP, and SPREAT using an appropriate dataset (e.g. "cami_X.pkl") with associated list of strings (e.g. "cami_text_X.pkl"), execute the following command:
python main.py --evaluate --soap --spreat --ctm --lda --cal-average --top-k 10 --X-name "cami_X.pkl" --text-name "cami_text_X.pkl" --vocab-name "biocyc_dictionary.pkl" --model-name "[model name (without prefix and extension)]" --mdpath "[path to the model]" --dspath "[path to the dataset]" --rspath "[path to store the results (e.g. costs)]" --batch 50 --num-epochs 10 --num-jobs 2
The same example applies for pretrained models (with collapse2ctm option) of SOAP and SPREAT.
To evaluate pretrained models (with --use-supplement and --use-features arguments) of SOAP and SPREAT using an appropriate dataset (e.g. "cami_X.pkl") with associated list of strings (e.g. "cami_text_X.pkl"), execute the following command:
python main.py --evaluate --soap --spreat --use-supplement --use-features --cal-average --top-k 10 --X-name "cami_X.pkl" --text-name "cami_text_X.pkl" --M-name "cami_M.pkl" --vocab-name "biocyc_dictionary.pkl" --features-name "biocyc_features.pkl" --model-name "[model name (without prefix and extension)]" --mdpath "[path to the model]" --dspath "[path to the dataset]" --rspath "[path to store the results (e.g. costs)]" --batch 50 --num-epochs 10 --num-jobs 2
To evaluate pretrained models of LDA, CTM, SOAP, and SPREAT using an text dataset (e.g. "pnas_Xtest.pkl") with associated list of strings (e.g. "pnas_text_Xtest.pkl"), execute the following command:
python main.py --evaluate --soap --spreat --ctm --lda --cal-average --top-k 10 --X-name "pnas_Xtest.pkl" --text-name "pnas_text_Xtest.pkl" --vocab-name "pnas_dictionary.pkl" --model-name "[model name (without prefix and extension)]" --mdpath "[path to the model]" --dspath "[path to the dataset]" --rspath "[path to store the results (e.g. costs)]" --batch 50 --num-epochs 10 --num-jobs 2
To transform data into components, we provide few examples.
Description about arguments in all examples: --batch is batch size, --num-jobs corresponds the number of parallel workers, --file-name is the name of file (without PREFIX and EXTENSION) of the transformed data, and --model-name corresponds the name of the learned model excluding PREFIX and EXTENSION. The model name will have .pkl extension. The argument --X-name is the input space of data (e.g. "cami_X.pkl" or "pnas_Xtest.pkl") while the argument --M-name corresponds the supplementary data (same size as X). Finally, --features-name corresponds the features of pathways obtained from pathway2vec.
The results of running all the commands below will be stored in --dspath which contains the transformed data.
To transform a dataset (e.g. "cami_X.pkl") using pretrained models of SOAP, SPREAT, CTM, and LDA, execute the following command:
python main.py --transform --soap --spreat --ctm --lda --X-name "cami_X.pkl" --model-name "[model name (without prefix and extension)]" --file-name "[the file name (without prefix and extension) of the transformed data]" --mdpath "[path to the model]" --dspath "[path to the dataset]" --batch 50 --num-jobs 2
The same example applies for pretrained models (with collapse2ctm option) of SOAP and SPREAT.
To transform a dataset (e.g. "cami_X.pkl") with supplementary pathways (e.g. "cami_M.pkl") using pretrained models (with --use-supplement and --use-features arguments) of SOAP and SPREAT, execute the following command:
python main.py --transform --soap --spreat --use-supplement --use-features --X-name "cami_X.pkl" --M-name "cami_M.pkl" --features-name "biocyc_features.pkl" --model-name "[model name (without prefix and extension)]" --file-name "[the file name (without prefix and extension) of the transformed data]" --mdpath "[path to the model]" --dspath "[path to the dataset]" --batch 50 --num-jobs 2
To transform a text dataset (e.g. "pnas_Xtest.pkl") using pretrained models of SOAP, SPREAT, CTM, and LDA, execute the following command:
python main.py --transform --soap --spreat --ctm --lda --X-name "pnas_Xtest.pkl" --model-name "[model name (without prefix and extension)]" --file-name "[the file name (without prefix and extension) of the transformed data]" --mdpath "[path to the model]" --dspath "[path to the dataset]" --batch 50 --num-jobs 2
If you find CHAP useful in your research, please consider citing the following paper:
- M. A. Basher, Abdur Rahman and Hallam, Steven J.. "Aggregating statistically correlated metabolic pathways into groups to improve pathway prediction outcomes.", arXiv (2022).
For any inquiries, please contact Steven Hallam and Abdurrahman Abul-Basher at: shallam@mail.ubc.ca and arbasher@student.ubc.ca