This project provides a script framework originally used to run benchmarks of BLAST and PLAST on a cluster infrastructure.
Provided scripts were originally designed to run comparison softwares on IFREMER's DATARMOR computer providing the PBS job scheduler system. However, it would be easy to adapt theses scripts with any other kind of job schedulers.
The basic principle of this benchmark was quite easy: find an optimal way to configure NCBI BLAST and INRIA PLAST sequence comparison tools in order to run them as efficiently as possible on a cluster computer. Tests were conducted by running bank-to-bank sequence comparisons of reasonnable sizes.
Our scripts were designed to work with:
In addition, you need some data sets to run sequence comparisons:
- FASTA files containing BLAST/PLAST queries
- BLAST databanks to be used as subjects for BLAST/PLAST comparison jobs
It is up to you to install all that material.
We used these scripts to evaluate the optimal configuration (cores, RAM and walltime) to run sequence comparison jobs (using either BLAST or PLAST) on the DATARMOR supercomputer available at IFREMER for bioinformatics computational intensive works.
For that purpose, we ran jobs using the following constraints:
- use 8, 16, 32 or 56 cores; DATARMOR provides several hundreds 56-core computing nodes;
- use 32 or 100Go RAM per computing node;
- queries: bacterium Yersinia pestis protein sequences (blastp and blastx) or gene sequences (blastn and megablast), i.e. 3979 sequences;
- subject banks: one of Swissprot (555,594 sequences), Genbank Bacteria division (1,604,589 sequences) or TrEmbl (90,050,708 sequences); banks content as available on September 2017.
- softwares: BLAST 2.2.31 & 2.6.0 and PLAST 2.3.2.
- BLAST cmd-line specific parameters: -max_target_seqs 1 -evalue 1e-3 -outfmt 9 (you can review the full BLAST cmd-line here; see end of script)
- PLAST cmd-line specific parameters: -max-hit-per-query 1 -max-hsp-per-hit 1 -e 1e-3 -outfmt 1 -seeds-use-ratio 0.01 (you can review the full PLAST cmd-line here; see end of script)
Below, we use the following identifiers to target each kind of queries:
- P: bacterium Yersinia pestis protein sequences
- N: bacterium Yersinia pestis gene sequences
Same for subject banks:
- P: SwissProt
- PL: TrEmbl
- N: Genbank Bacteria division (to be used to run blastn)
- M: Genbank Bacteria division (to be used to run megablast)
Using these query and subject naming codes, we introduce the following jobs names:
- P-P: a protein-protein comparison of bacterium Yersinia pestis protein sequences against SwissProt
- P-PL: a protein-protein comparison of bacterium Yersinia pestis protein sequences against TrEmbl
- N-N: a nucleotide-nucleotide (regular BLASTn) comparison of bacterium Yersinia pestis gene sequences against Genbank Bacteria division
- M-N: a nucleotide-nucleotide (M egablast) comparison of bacterium Yersinia pestis gene sequences against Genbank Bacteria division
- N-P: a nucleotide-protein comparison of bacterium Yersinia pestis gene sequences against SwissProt
- N-PL: a nucleotide-protein comparison of bacterium Yersinia pestis gene sequences against TrEMBL
Raw results (running times, CPU & memory use on 8, 16 ,32 and 56 cores) are as follows:
Note: content of the above files is described here
Since we were interested in running times, here are some graphical outputs (generated using material from sub-folder gnuplot):
-
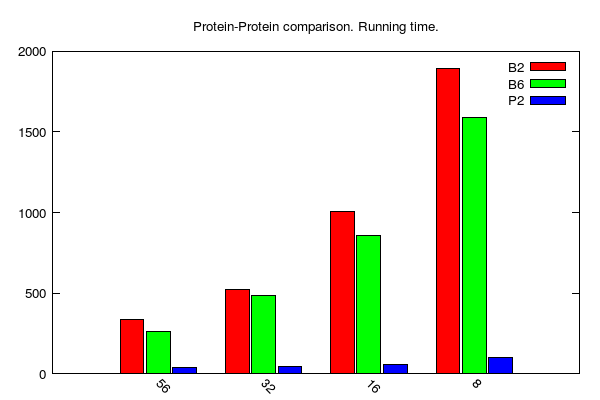
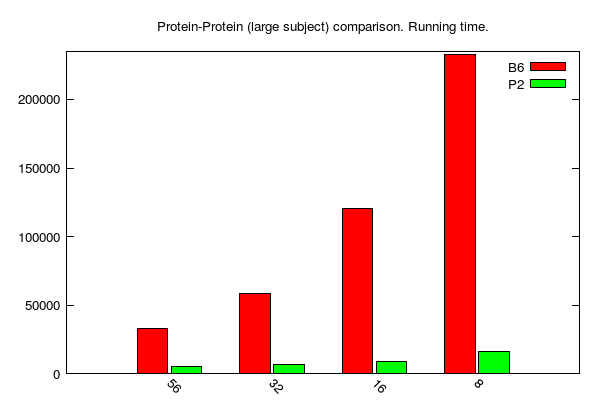
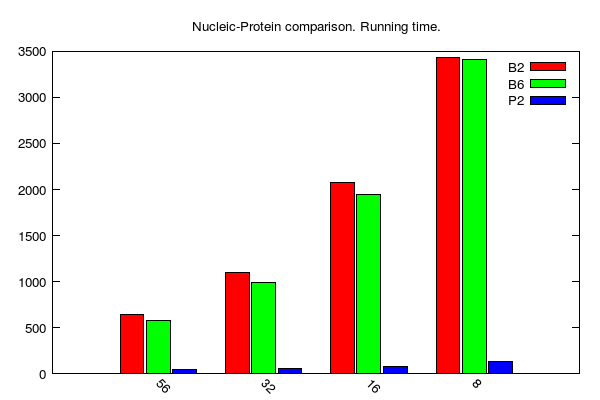
y-axis: running time (seconds)
-
x-axis: number of cores
-
B2, B6 and P stands for BLAST 2.2.31, BLAST 2.6.0 and PLAST 2.3.2, respectively
-
BLASTp/PLASTp medium jobs (data table is here):
-
BLASTp/PLASTp large jobs (data table is here):
-
BLASTx/PLASTx jobs (data table is here):
-
BLASTn jobs (data table is here):
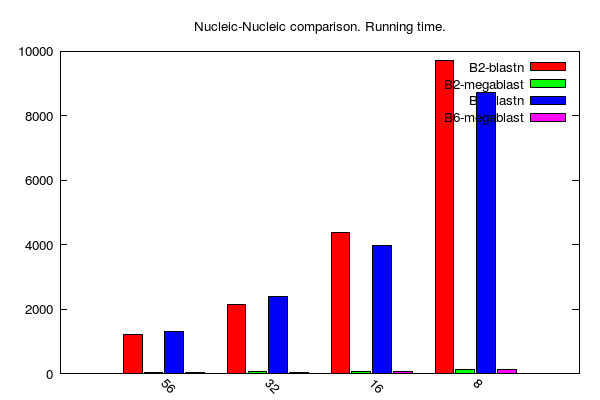
-
Megablast jobs (data table is here):
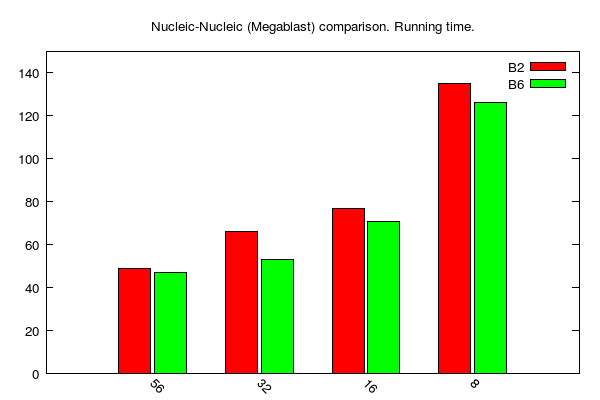
Note: N-N and M-N comparisons not done using PLAST since it is not optimal with regards to BLAST performance.
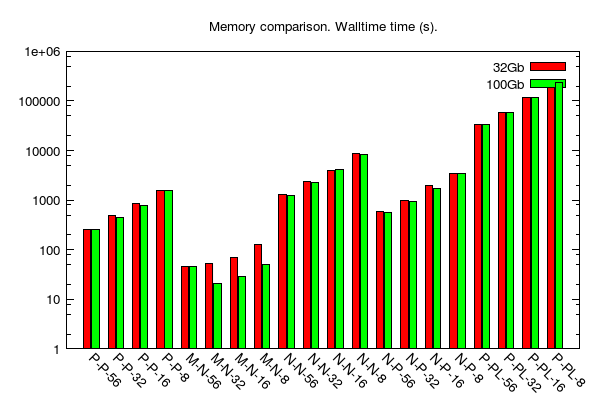
Here, we were interested to check whether or not BLAST running times change given the amount of RAM available on a computing node. We used two configurations: 32Gb vs. 100Gb (all other search parameters were the same over the various jobs).
-
- y-axis: log scale of running time (i.e. walltime); unit is seconds.
- x-axis: type of comparison; e.g. "P-P-56" = protein-protein comparison on 56 cores.
- data table is here
In this section, we describe the use of HTC-BLAST 4.3 framework to dispatch BLAST jobs.
For these tests, we dispatched BLAST jobs on one up to five 28-core nodes on DATARMOR's machine.
We used the same data sets (i.e. query and subject banks) as for BLAST/PLAST tests, above presented.
On the following plots:
-
y-axis: running time (i.e. walltime); unit is seconds
-
x-axis: number of cores (28: one 28-core computing node; 56: two 28-core computing nodes; 84: three 28-core computing nodes; and so on...)
-
HTC-BLASTp jobs (data table is here):
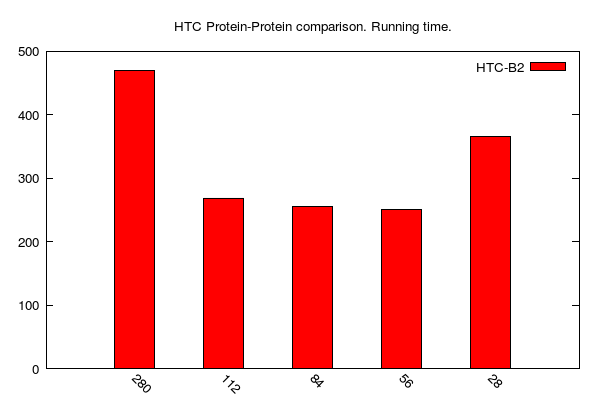
-
HTC-BLASTx jobs (data table is here):
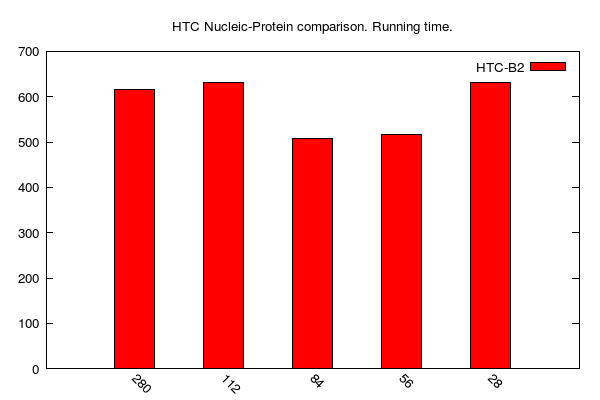
-
HTC-BLASTn jobs (data table is here):
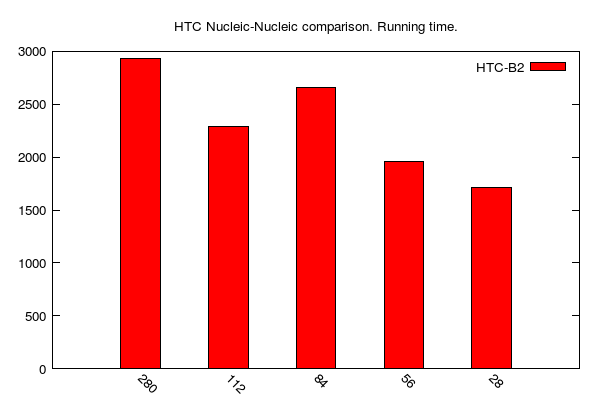
-
HTC-Megablast jobs (data table is here):
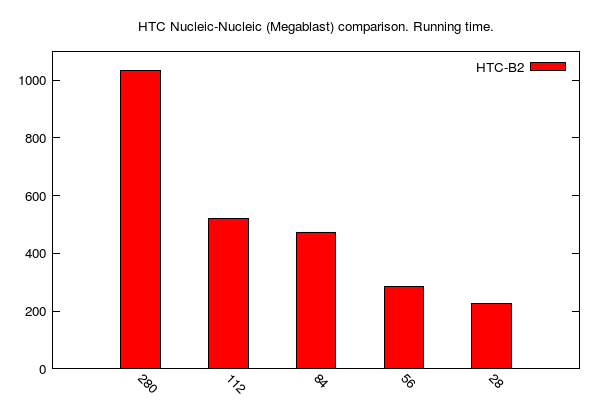
Since we used the same data sets as above, we can appreciate (or not) the advantages of HTC-BLAST over regular BLAST to accelerate (or not) sequence comparisons. In the following table, we compare job running times (i.e. "walltime") of HTC vs Regular BLAST on 56 cores:
| Job kind | Regular BLAST | HTC-BLAST |
|---|---|---|
| P-P | 338 s | 251 s |
| N-P | 647 s | 517 s |
| N-N | 1226 s | 1985 s |
| M-N | 49 s | 286 s |
Regular NCBI BLAST software provides a remote feature that enables a user to run a BLAST job using NCBI computational infrastructure.
To check whether or not it could be of interest to use NCBI or local IT resources, we conducted a single test (NCBI being a public shared resource, we did not run many tests).
Our test was as simple as using NCBI's web_blast.pl script to run our "P-P" comparison (see previous sections):
time ./web_blast.pl blastp swissprot Genbank_Yersinia_Angola_prot00
RID is: 58VWKSPB015
Search complete, retrieving results...
done.
real 184m32,700s
user 0m35,217s
sys 0m1,444s
As one can see, such a job has taken 184 minutes at NCBI (data transfer + comparison) instead of less than 5 minutes on a 56-core DATARMOR's computing node (check out this table for "P-P" runtimes on 8, 16 and 32 cores).