Recently, there has been an ascension of workflow languages to create reproducible, cloud computing agnostic, parallizable, and provenence tracked pipelines.
- Stallman, R.M. and McGrath, R., 1991. GNU Make-A Program for Directing Recompilation. note: appeared in 1976, this links to the GNU Make manual.
- Köster, J. and Rahmann, S., 2012. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics, 28(19), pp.2520-2522.
- Amstutz, P., Tijanić, N., Soiland-Reyes, S., Kern, J., Stojanovic, L., Pierce, T., Chilton, J., Mikheev, M., Lampa, S., Ménager, H. and Frazer, S., 2015, July. Portable workflow and tool descriptions with the CWL. In Bioinformatics Open Source Conference.
- Amstutz, P., Crusoe, M.R., Tijanić, N., Chapman, B., Chilton, J., Heuer, M., Kartashov, A., Leehr, D., Ménager, H., Nedeljkovich, M. and Scales, M., 2016. Common workflow language, v1.0.
- Di Tommaso, P., Chatzou, M., Floden, E.W., Barja, P.P., Palumbo, E. and Notredame, C., 2017. Nextflow enables reproducible computational workflows. Nature biotechnology, 35(4), pp.316-319.
- Michael Kotliar, Andrey V Kartashov, Artem Barski, CWL-Airflow: a lightweight pipeline manager supporting Common Workflow Language, GigaScience, Volume 8, Issue 7, July 2019, giz084, https://doi.org/10.1093/gigascience/giz084
- Yuen, D., Cabansay, L., Duncan, A., Luu, G., Hogue, G., Overbeck, C., Perez, N., Shands, W., Steinberg, D., Reid, C. and Olunwa, N., 2021. The Dockstore: enhancing a community platform for sharing reproducible and accessible computational protocols. Nucleic acids research, 49(W1), pp.W624-W632.
- Gorzalski, A.J., Boyles, C., Sepcic, V., Verma, S., Sevinsky, J., Libuit, K., Van Hooser, S. and Pandori, M.W., 2022. Rapid repeat infection of SARS-CoV-2 by two highly distinct delta-lineage viruses. Diagnostic Microbiology and Infectious Disease, 104(1), p.115747.
Here we aim to explore, compare, and contrast several workflow languages for our internal workflow development. There are several academic papers that explore these workflow languages in certain domains, especially relating to bioinformatic analysis.
A Basic makefile
all:
echo "hello world"
From commandline
make
#> echo "Hello World"
#> Hello World
Snakemake is a python program and can be installed via miniconda using the provided environment.yml file.
conda env create -f environment.yml
conda activate workflow_env
Create snake_say_hi.txt
rule all:
input: "hello_world.txt"
rule hello_world:
output: "hello_world.txt"
shell: "echo Hello World > hello_world.txt"
It looks basically like a Makefile. Run it via:
snakemake --snakefile snake_say_hi.txt -j1
which gives:
Building DAG of jobs...
Using shell: /bin/bash
Provided cores: 1 (use --cores to define parallelism)
Rules claiming more threads will be scaled down.
Job counts:
count jobs
1 all
1 hello_world
2
Select jobs to execute...
[Wed Feb 3 17:49:58 2021]
rule hello_world:
output: hello_world.txt
jobid: 1
[Wed Feb 3 17:49:58 2021]
Finished job 1.
1 of 2 steps (50%) done
Select jobs to execute...
[Wed Feb 3 17:49:58 2021]
localrule all:
input: hello_world.txt
jobid: 0
[Wed Feb 3 17:49:58 2021]
Finished job 0.
2 of 2 steps (100%) done
Complete log: /Users/jenchang/Desktop/2021-02-03_Snakemake/test_snakemake/.snakemake/log/2021-02-03T174958.315655.snakemake.log
Still in development
- install via miniconda (based on github: cwl).
name: cwl_env
channels:
- conda-forge
- bioconda
- defaults
dependencies:
- python=3.8
- cwltool
- run
conda env create -f cwl_env.yml
CWL needs two files, a cwl and a process file... in progress
conda activate cwl_env
cwltool --version # <= check if it works, other documentation seems to call this cwl-runner...
... copy from other repo...
We created a minimal nextflow pipeline example early in 2020. This will be modified fof snakemake and CWL.
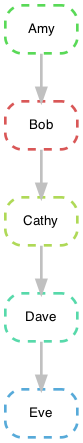
We define 5 processes (nextflow) or rules (snakemake) that pass a text file from Amy to Eve.
Amy -> Bob -> Cathy -> Dave -> Eve
Copied from https://github.com/j23414/desc_workflows
Instead of worrying about installing a long running program, we’ll
simulate it using the sleep 5 command (wait 5 seconds).
Create a textfile pass_baton.nf, the nf extention indicates a nextflow script.
#! /usr/bin/env nextflow
/**********************************
Create a chain of long running processes
This basically simulates trinity/canu/whatever pipeline
**********************************/
println "\nPipeline = Amy -> Bob -> Cathy -> Dave -> Eve"
println " where each person runs 5 seconds to pass the baton to next person\n"
process Amy {
output: stdout Amy_out
script:
"""
#! /usr/bin/env bash
sleep 5 # <= pause for a few seconds
echo "Amy passes baton"
"""
}
process Bob {
input: val baton_in from Amy_out
output: stdout Bob_out
script:
"""
#! /usr/bin/env bash
sleep 5 # <= pause for a few seconds
echo "$baton_in; Bob passes baton"
"""
}
process Cathy {
input: val baton_in from Bob_out
output: stdout Cathy_out
script:
"""
#! /usr/bin/env bash
sleep 5 # <= pause for a few seconds
echo "$baton_in; Cathy passes baton"
"""
}
process Dave {
input: val baton_in from Cathy_out
output: stdout Dave_out
script:
"""
#! /usr/bin/env bash
sleep 5 # <= pause for a few seconds
echo "$baton_in; Dave passes baton"
"""
}
process Eve {
input: val baton_in from Dave_out
output: stdout Eve_out
script:
"""
#! /usr/bin/env bash
sleep 5 # <= pause for a few seconds
echo "$baton_in; Eve passes baton"
"""
}
println Eve_out.view { it.trim() }
Which looks nice in bash… as it prints progress
N E X T F L O W ~ version 20.04.1
Launching `code/script06.nf` [crazy_mclean] - revision: c6a509673f
Pipeline = Amy -> Bob -> Cathy -> Dave -> Eve
where each person runs 5 seconds to pass the baton to next person
DataflowVariable(value=null)
executor > local (2)
[ee/41b22c] process > Amy [100%] 1 of 1 ✔
[cf/db02ae] process > Bob [ 0%] 0 of 1
[- ] process > Cathy -
[- ] process > Dave -
[- ] process > Eve -
Eventually looks like the following when finished:
nextflow run script06.nf
N E X T F L O W ~ version 20.04.1
Launching `code/script06.nf` [crazy_mclean] - revision: c6a509673f
Pipeline = Amy -> Bob -> Cathy -> Dave -> Eve
where each person runs 5 seconds to pass the baton to next person
DataflowVariable(value=null)
executor > local (5)
[ee/41b22c] process > Amy [100%] 1 of 1 ✔
[cf/db02ae] process > Bob [100%] 1 of 1 ✔
[b0/cccd94] process > Cathy [100%] 1 of 1 ✔
[9f/a652c6] process > Dave [100%] 1 of 1 ✔
[ca/39a72a] process > Eve [100%] 1 of 1 ✔
Amy passes baton
; Bob passes baton
; Cathy passes baton
; Dave passes baton
; Eve passes baton
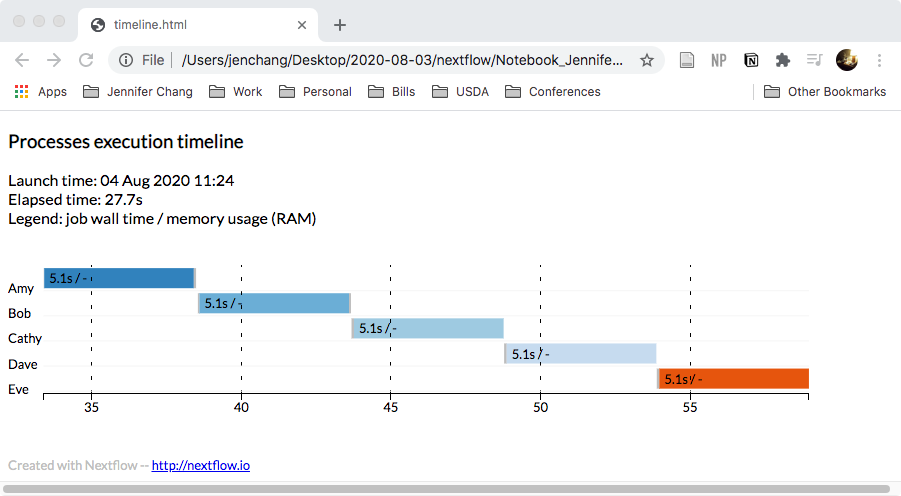
And if you run it with the html reports turned on:
nextflow run script06.nf -with-timeline timeline.html
It will also generate a plot:
Snakefile <- best practice, ah how do deal wth multiple snakemake pipelines in one folder?
rule all:
input: "Eve_baton.txt"
rule Eve:
input: "Dave_baton.txt"
output: "Eve_baton.txt"
shell: "sleep 5; cat {input} > {output}; echo 'Eve passes baton' >> {output};"
rule Dave:
input: "Cathy_baton.txt"
output: "Dave_baton.txt"
shell: "sleep 5; cat {input} > {output}; echo 'Dave passes baton' >> {output};"
rule Cathy:
input: "Bob_baton.txt"
output: "Cathy_baton.txt"
shell: "sleep 5; cat {input} > {output}; echo 'Cathy passes baton' >> {output};"
rule Bob:
input: "Amy_baton.txt"
output: "Bob_baton.txt"
shell: "sleep 5; cat {input} > {output}; echo 'Bob passes baton' >> {output};"
rule Amy:
output: "Amy_baton.txt"
shell: "sleep 5; echo 'Amy passes baton' > {output}"
Run it below:
snakemake -j1 # <= will automatically look for a snakefile
Building DAG of jobs...
Using shell: /bin/bash
Provided cores: 1 (use --cores to define parallelism)
Rules claiming more threads will be scaled down.
Job counts:
count jobs
1 Amy
1 Bob
1 Cathy
1 Dave
1 Eve
1 all
6
Select jobs to execute...
[Tue Feb 23 17:47:02 2021]
rule Amy:
output: Amy_baton.txt
jobid: 5
[Tue Feb 23 17:47:03 2021]
Finished job 5.
1 of 6 steps (17%) done
Select jobs to execute...
[Tue Feb 23 17:47:03 2021]
rule Bob:
input: Amy_baton.txt
output: Bob_baton.txt
jobid: 4
[Tue Feb 23 17:47:03 2021]
Finished job 4.
2 of 6 steps (33%) done
Select jobs to execute...
[Tue Feb 23 17:47:03 2021]
rule Cathy:
input: Bob_baton.txt
output: Cathy_baton.txt
jobid: 3
[Tue Feb 23 17:47:03 2021]
Finished job 3.
3 of 6 steps (50%) done
Select jobs to execute...
[Tue Feb 23 17:47:03 2021]
rule Dave_baton:
input: Cathy_baton.txt
output: Dave_baton.txt
jobid: 2
[Tue Feb 23 17:47:03 2021]
Finished job 2.
4 of 6 steps (67%) done
Select jobs to execute...
[Tue Feb 23 17:47:03 2021]
rule Eve_baton:
input: Dave_baton.txt
output: Eve_baton.txt
jobid: 1
[Tue Feb 23 17:47:03 2021]
Finished job 1.
5 of 6 steps (83%) done
Select jobs to execute...
[Tue Feb 23 17:47:03 2021]
localrule all:
input: Eve_baton.txt
jobid: 0
[Tue Feb 23 17:47:03 2021]
Finished job 0.
6 of 6 steps (100%) done
Complete log: /Users/jenchang/Desktop/ttt/test_snakemake/.snakemake/log/2021-02-23T174702.949455.snakemake.log
less Eve_baton.txt
#> Amy passes baton
#> Bob passes baton
#> Cathy passes baton
#> Dave passes baton
#> Eve passes baton
Maybe there's a similar way to print out a timeline in Snakemake? Ah, found it.
snakemake --dag Eve_baton.txt --snakefile snakemake | dot -Tpng > dag.png
[ ] Urmi suggested exploring snakemake html reports:
- Manual - https://snakemake.readthedocs.io/en/stable/snakefiles/reporting.html
- Example - https://koesterlab.github.io/resources/report.html
snakemake --report report.html
Create next_baton.nf in DSL2 style
#! /usr/bin/env nextflow
nextflow.enable.dsl=2
/**********************************
Create a chain of long running processes
This basically simulates trinity/canu/whatever pipeline
**********************************/
println "\nPipeline = Amy -> Bob -> Cathy -> Dave -> Eve"
println " where each person runs 5 seconds to pass the baton to next person\n"
process Amy {
output: stdout
script:
"""
#! /usr/bin/env bash
sleep 5 # <= pause for a few seconds
echo "Amy passes baton"
"""
}
process Bob {
input: val baton_in
output: stdout
script:
"""
#! /usr/bin/env bash
sleep 5 # <= pause for a few seconds
echo "$baton_in; Bob passes baton"
"""
}
process Cathy {
input: val baton_in
output: stdout
script:
"""
#! /usr/bin/env bash
sleep 5 # <= pause for a few seconds
echo "$baton_in; Cathy passes baton"
"""
}
process Dave {
input: val baton_in
output: stdout
script:
"""
#! /usr/bin/env bash
sleep 5 # <= pause for a few seconds
echo "$baton_in; Dave passes baton"
"""
}
process Eve {
input: val baton_in
output: stdout
script:
"""
#! /usr/bin/env bash
sleep 5 # <= pause for a few seconds
echo "$baton_in; Eve passes baton"
"""
}
/* === Main workflow */
workflow {
Amy | Bob | Cathy | Dave | Eve | view
}
Run next_baton.nf, notice section workflow... this is the main addition of DSL2. We could reorganize that pipeline to it went from Amy | Dave | Bob | Eve | view, etc. In the original DSL1, inputs were labled, so we couldn't change order.
nextflow run next_baton.nf
N E X T F L O W ~ version 20.07.1
Launching `next_baton.nf` [sharp_agnesi] - revision: 6c35007a22
Pipeline = Amy -> Bob -> Cathy -> Dave -> Eve
where each person runs 5 seconds to pass the baton to next person
executor > local (5)
[64/1713d1] process > Amy [100%] 1 of 1 ✔
[10/c670da] process > Bob [100%] 1 of 1 ✔
[f2/4d2032] process > Cathy [100%] 1 of 1 ✔
[91/c60628] process > Dave [100%] 1 of 1 ✔
[d7/61db2e] process > Eve [100%] 1 of 1 ✔
Amy passes baton
; Bob passes baton
; Cathy passes baton
; Dave passes baton
; Eve passes baton
Makefile
all: Eve_baton.txt
cat Eve_baton.txt
Eve_baton.txt: Dave_baton.txt
cat Dave_baton.txt > Eve_baton.txt
echo "Eve passes the baton" >> Eve_baton.txt
Dave_baton.txt: Cathy_baton.txt
cat Cathy_baton.txt > Dave_baton.txt
echo "Dave passes the baton" >> Dave_baton.txt
Cathy_baton.txt: Bob_baton.txt
cat Bob_baton.txt > Cathy_baton.txt
echo "Cathy passes the baton" >> Cathy_baton.txt
Bob_baton.txt: Amy_baton.txt
cat Amy_baton.txt > Bob_baton.txt
echo "Bob passes the baton" >> Bob_baton.txt
Amy_baton.txt:
echo "Amy passes the baton" > Amy_baton.txt
In bash, run "make"
make
#> echo "Amy passes the baton" > Amy_baton.txt
#> cat Amy_baton.txt > Bob_baton.txt
#> echo "Bob passes the baton" >> Bob_baton.txt
#> cat Bob_baton.txt > Cathy_baton.txt
#> echo "Cathy passes the baton" >> Cathy_baton.txt
#> cat Cathy_baton.txt > Dave_baton.txt
#> echo "Dave passes the baton" >> Dave_baton.txt
#> cat Dave_baton.txt > Eve_baton.txt
#> echo "Eve passes the baton" >> Eve_baton.txt
#> cat Eve_baton.txt
#> Amy passes the baton
#> Bob passes the baton
#> Cathy passes the baton
#> Dave passes the baton
#> Eve passes the baton