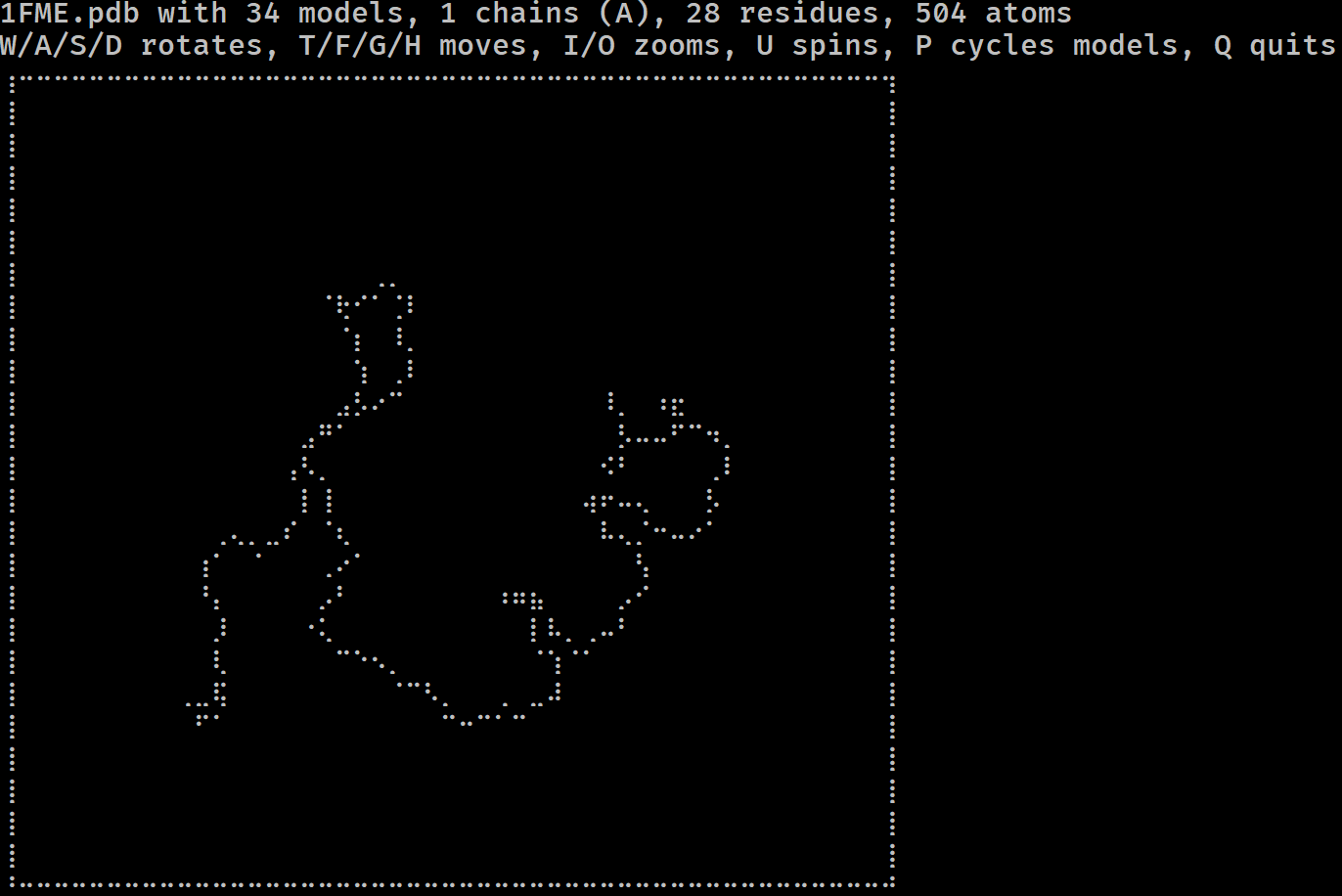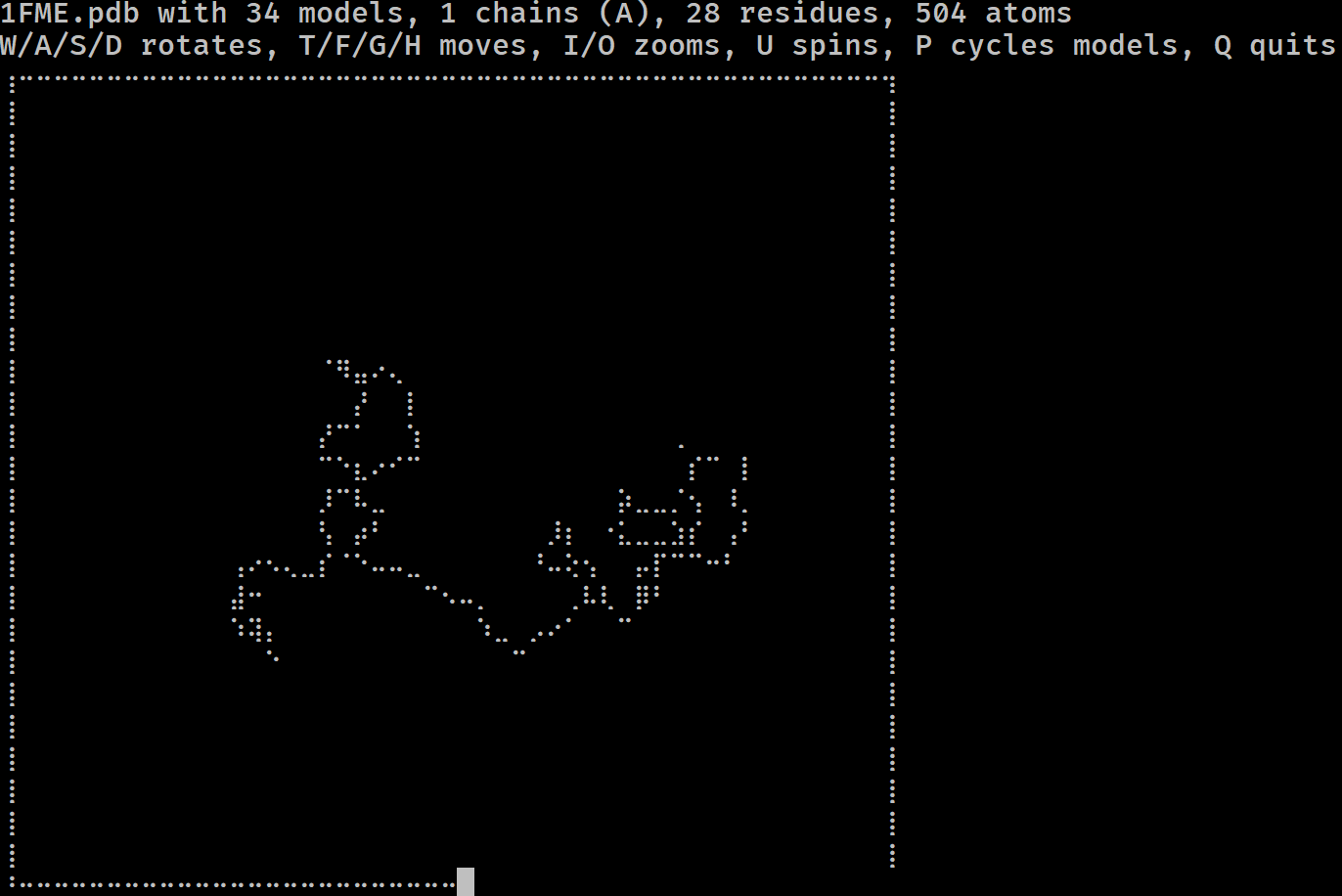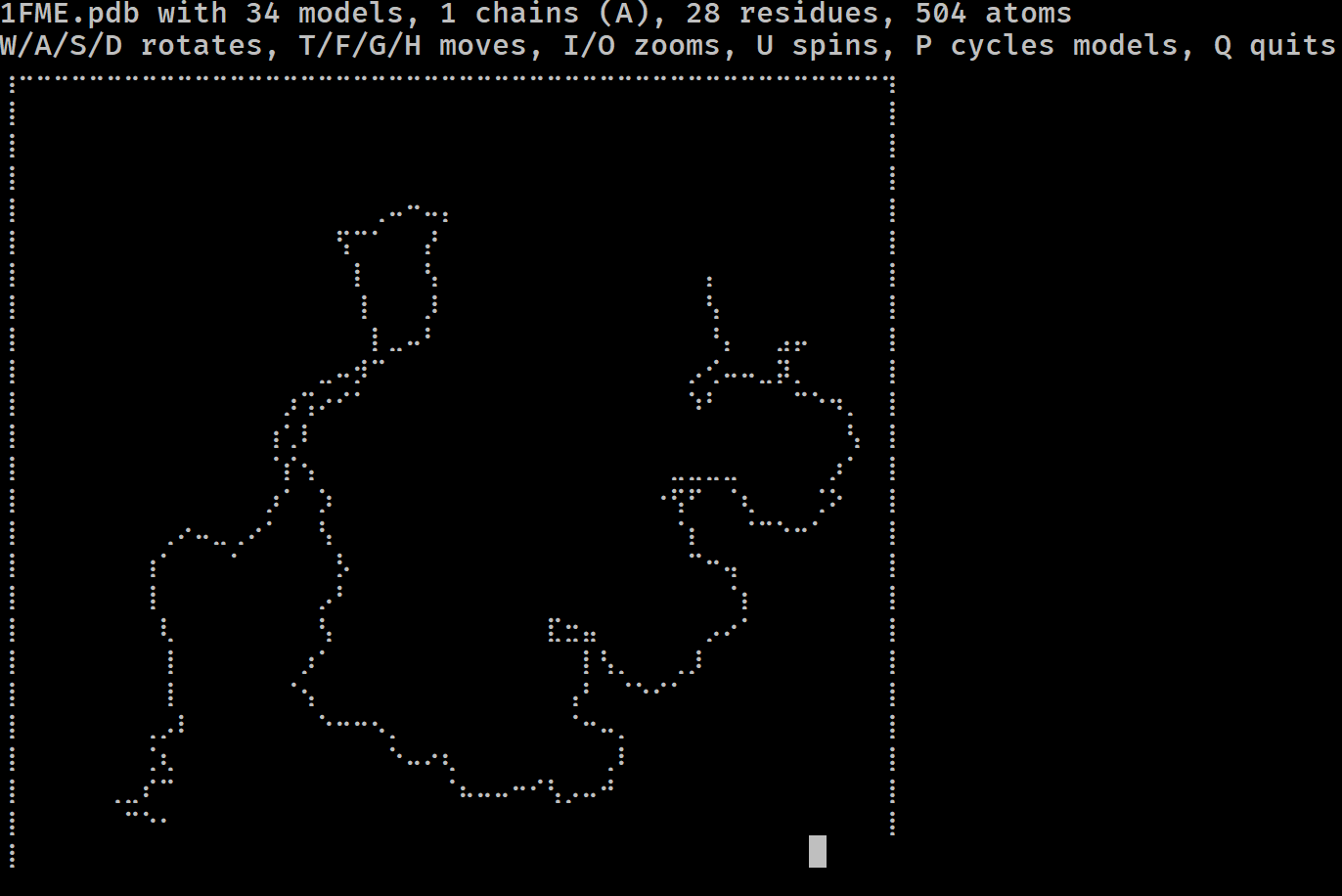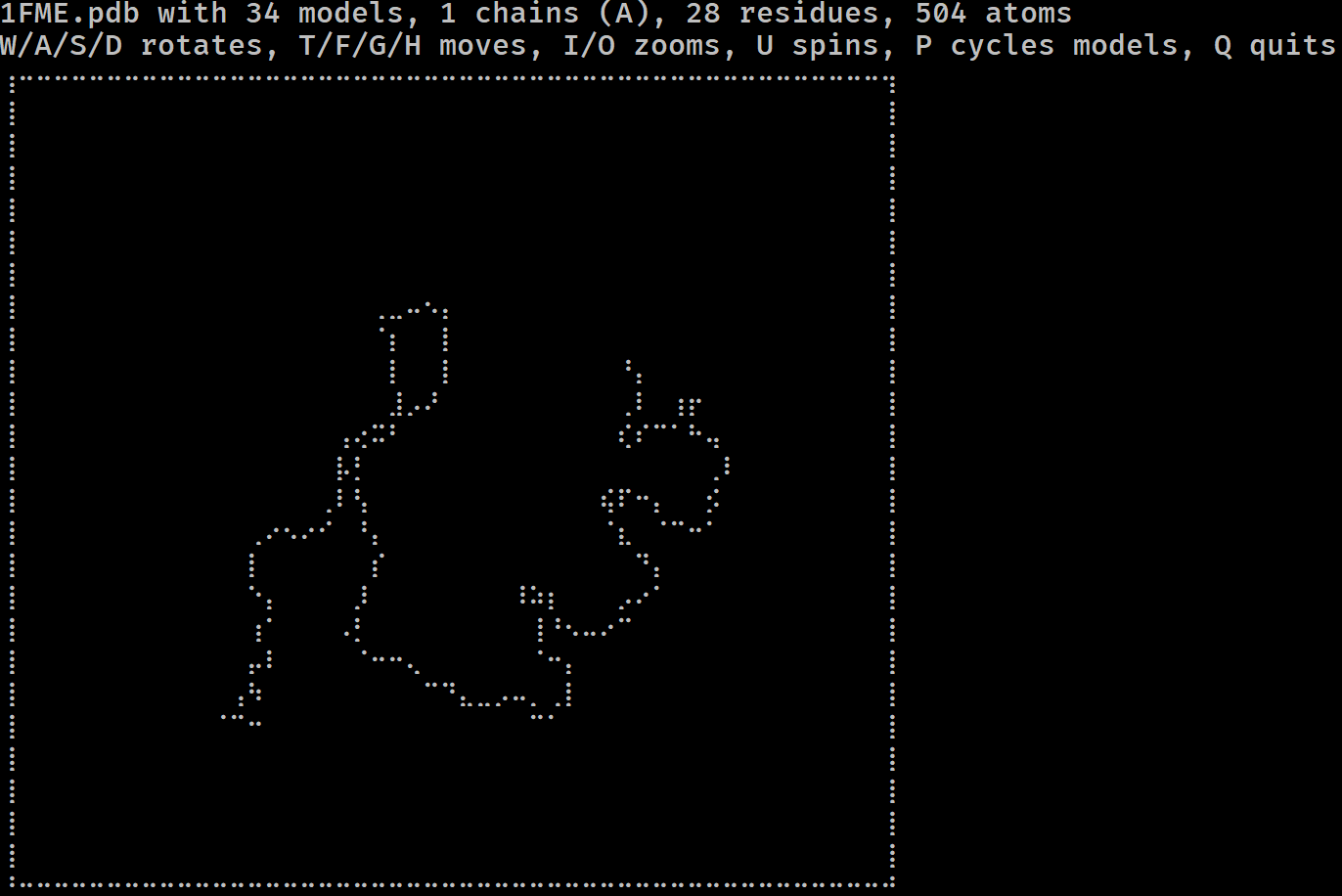
Have you ever wondered if you could visualise a protein in the terminal? Do you miss the resolution of 1990s RasMol? More seriously, have you ever wanted to get a feel for a protein/DNA/RNA structure on a remote system without having to copy it locally and load it in PyMol?
If so, mmterm is for you. It gives you a quick interactive view of a PDB/mmCIF/MMTF/MAE/MAEGZ file, purely in the terminal. It should work anywhere that the Python curses module works, i.e. anywhere but Windows. It is built on top of drawille, NumPy, Biopython and Schrödinger.
Python 3 is required.
git clone https://github.com/jgreener64/mmterm
cd mmterm
pip install -e .Installation puts the executable mmterm on the path.
Run mmterm -h to see the help text.
To view a structure, just give the filepath:
mmterm 1FME.pdbControls for rotating, translating, zooming, spinning and playing trajectories are given.
The file format is guessed from the file extension but can be given explicitly with -f:
mmterm 1FME -f mmcifOther options:
# Show a different model initially
mmterm 1FME.pdb -m 5
# Only show given chains
mmterm 4KPY.pdb -c A B
# Decrease the size of the viewing box
mmterm 1FME.pdb -s 50
# Read from stdin
cat 1FME.pdb | mmterm -f pdb -You can also use mmterm to view proteins from the REPL, from IPython or from other Python scripts:
from mmterm import view
view("1FME.pdb")