End-to-End Automated Medical Image Analysis on the UCSF Clinical PACS
John Colby MD PhD, Jeffrey Rudie MD PhD, Andreas Rauschecker MD PhD, Leo Sugrue MD PhD, Janine Lupo PhD, Jason Crane PhD, Christopher Hess MD PhD, Javier Villanueva-Meyer MD
ASNR 2020 annual meeting (oral presentation #1717)
- Slides are hosted on github.io: https://johncolby.github.io/asnr2020
- Video presentation is hosted on vimeo: https://vimeo.com/419503226 (also embedded in the slideshow, above)
- PDF snapshots of the slides are included in this repository: asnr2020.pdf
- ePoster is also publicly available on the conference website: American Society of Neuroradiology (ASNR) 58th Annual Meeting 2020
rad_apps- Application framework for analysis on the radiology clinical PACS.dcmclass- Tools for identifying imaging modality based on DICOM header data.rsna_heme- Automated detection and classification of intracranial hemorrhage at noncontrast CT.brats_preprocessing- Tools for preprocessing clinical data according to the BraTS specification.unet_brats- 3D U-Net deep learning segmentation.
The availability of state of the art algorithms for analyzing spatial image data, and the high performance computing resources needed to train them, are quickly reaching the point of commoditization. Therefore, increasing relative importance is shifting to the training data/annotations needed to drive these models, and to the engineering solutions needed to deploy these models as real-world products at the point of care.
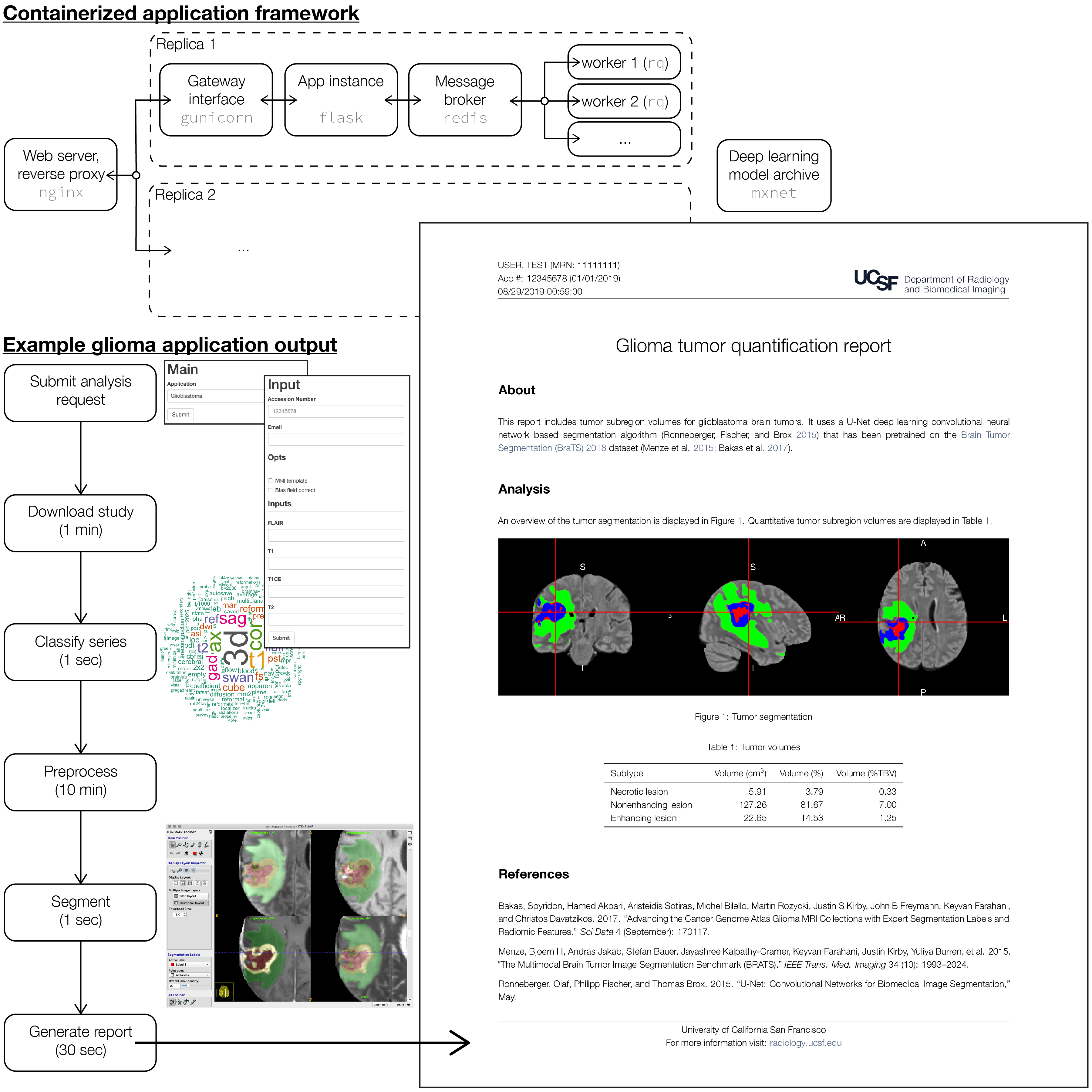
Here we present engineering development work on an end-to-end platform for automated image analysis on our clinical PACS. A simple extensible web application front end is used to enter study accession number (hookable from PACS), choose application (e.g. glioma segmentation, metastasis detection, etc.), prescribe analysis options, and submit the analysis request. Once study data are extracted from PACS, machine learning tools are used to identify the desired series based on fitting an application-specific ensembled tree model (e.g. gradient boosted trees or random forest) to the DICOM header data. Raw imaging data are preprocessed using best practices for data parallel pipelining, provenance, and reproducible research (i.e. nipype). An inference model server is used to expose a variety of custom pre-trained deep learning models through a common application programming interface. Results (e.g. segmentation label maps for the glioma context) are post-processed to generate structured data ready for warehousing, and stored for potential tune up and retraining on problem cases. Finally, using R Markdown and a customized LaTeX report template, we generate automated dynamic analysis reports according to our institutional brand graphic identity (for automated delivery to the referring clinician, or for upload back into the EMR or PACS). Importantly, these components are fully containerized, and deployed as a scalable production-ready cluster via the docker swarm orchestration framework.
The schematic of this application framework, together with representative output from the glioma segmentation context, is displayed in Figure 1.
Using open source software tools and commodity hardware, this project demonstrates the broad accessibility with which state of the art methods for medical image analysis may be integrated into the clinical radiology enterprise.