Code for Modeling Dense Multimodal Interactions Between Biological Pathways and Histology for Survival Prediction
Welcome to the official GitHub repository for our CVPR 2024 paper, "Modeling Dense Multimodal Interactions Between Biological Pathways and Histology for Survival Prediction". This project was developed by the Mahmood Lab at Harvard Medical School and Brigham and Women's Hospital. Preprint can be accessed here.
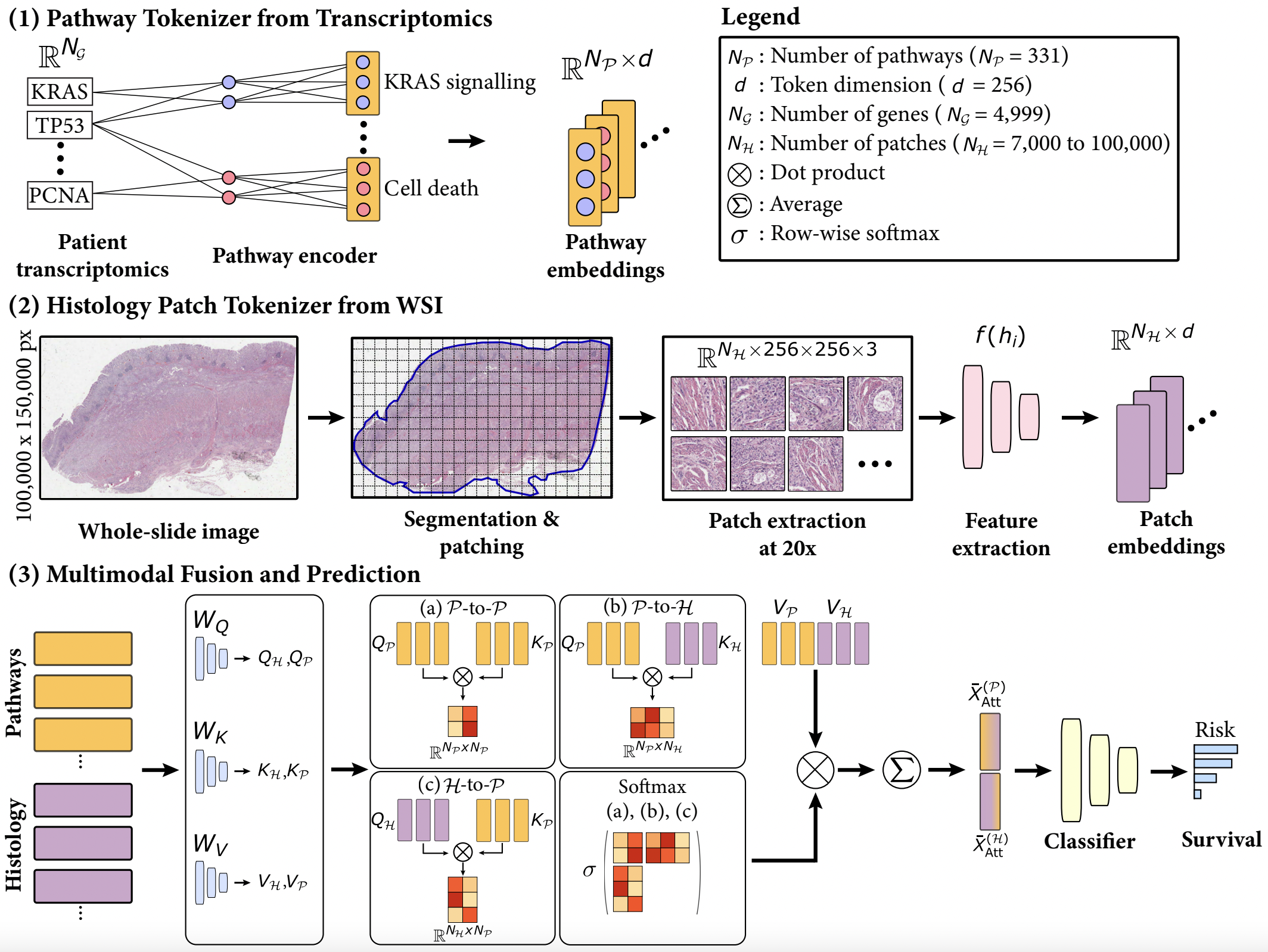
In our study, we explore the integration of whole-slide images (WSIs) and bulk transcriptomics to enhance patient survival prediction and interpretability. We focus on addressing two key challenges: (1) devising a method for meaningful tokenization of transcriptomics data and (2) capturing dense multimodal interactions between WSIs and transcriptomics. Our proposed model, SurvPath, leverages biological pathway tokens from transcriptomics and histology patch tokens from WSIs, facilitating memory-effective fusion through a multimodal Transformer. SurvPath surpasses unimodal and multimodal baselines across five datasets from The Cancer Genome Atlas, showcasing state-of-the-art performance. Furthermore, our interpretability framework identifies critical multimodal prognostic factors, offering deeper insights into genotype-phenotype interactions and underlying biological mechanisms.
- Linux (Tested on Ubuntu 18.04)
- NVIDIA GPU (Tested on Nvidia GeForce RTX 2080 Ti x 16) with CUDA 11.0 and cuDNN 7.5
- Python (3.8.13), h5py (2.10.0), matplotlib (3.6.3), numpy (1.21.6), opencv-python (4.5.1.48), openslide-python (1.2.0), openslide (3.4.1), pandas (1.4.2), pillow (9.0.1), PyTorch (1.6.5), scikit-learn (1.2.1), scipy (1.9.0), torchvision (0.13.1), captum (0.6.0), shap (0.41.0)
To download diagnostic WSIs (formatted as .svs files), molecular feature data and other clinical metadata, please refer to the NIH Genomic Data Commons Data Portaland the cBioPortal. WSIs for each cancer type can be downloaded using the GDC Data Transfer Tool. To get the pathway compositions for 50 Hallmarks, refer to MsigDB. To get the Reactome pathway compositions, refer to PARADIGM
To process Whole Slide Images (WSIs), first, the tissue regions in each biopsy slide are segmented using Otsu's Segmentation on a downsampled WSI using OpenSlide. The 256 x 256 patches without spatial overlapping are extracted from the segmented tissue regions at the desired magnification. Consequently, an SSL pretrained Swin Transformer CTransPath is used to encode raw image patches into 768-dim feature vectors, which we then save as .pt files for each WSI. The extracted features then serve as input (in a .pt file) to the network. All pre-processing of WSIs is done using the CLAM toolbox.
We downloaded raw RNA-seq abundance data for the TCGA cohorts from the Xena database and performed normalization in the dataset class. The raw data is included as CSV files datasets_csv. Xena database was also used to access disease specific survival and associated censorhsip. Using the Reactome and MSigDB Hallmarks pathway compositions, we selected pathways that had more than 90% of transcriptomics data available. The compositions can be found at metadata.
For evaluating the algorithm's performance, we partitioned each dataset using 5-fold cross-validation (stratified by the site of histology slide collection). Splits for each cancer type are found in the splits folder, which each contain splits_{k}.csv for k = 1 to 5. In each splits_{k}.csv, the first column corresponds to the TCGA Case IDs used for training, and the second column corresponds to the TCGA Case IDs used for validation. Slides from one case are not distributed across training and validation sets. Alternatively, one could define their own splits, however, the files would need to be defined in this format. The dataset loader for using these train-val splits are defined in the return_splits function in the SurvivalDatasetFactory.
Refer to scripts folder for source files to train SurvPath and the baselines presented in the paper. Refer to the paper to find the hyperparameters required for training.
- The preferred mode of communication is via GitHub issues.
- If GitHub issues are inappropriate, email
avaidya@mit.edu(and ccgjaume@bwh.harvard.edu). - Immediate response to minor issues may not be available.
If you find our work useful in your research, please consider citing our paper at:
@article{jaume2023modeling,
title={Modeling Dense Multimodal Interactions Between Biological Pathways and Histology for Survival Prediction},
author={Jaume, Guillaume and Vaidya, Anurag and Chen, Richard and Williamson, Drew and Liang, Paul and Mahmood, Faisal},
journal={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)},
year={2024}
}
Mahmood Lab - This code is made available under the GPLv3 License and is available for non-commercial academic purposes.