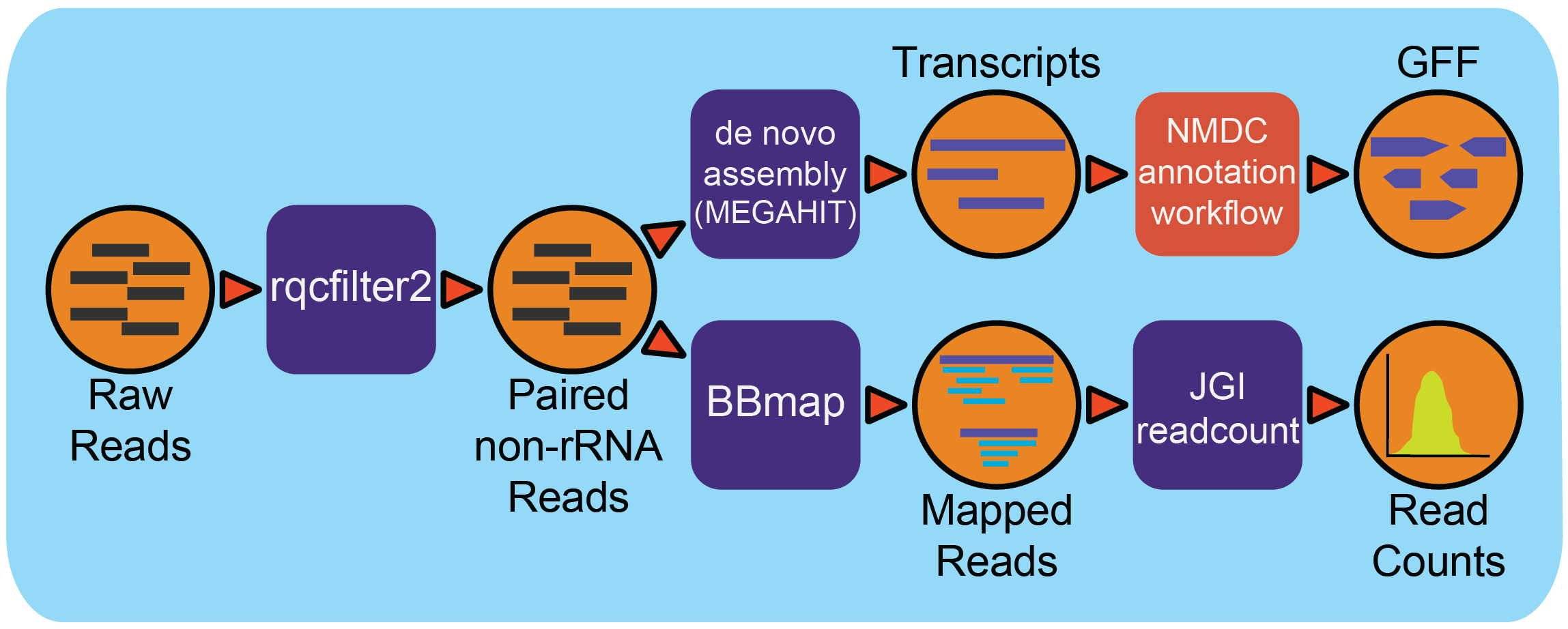
This workflow is designed to analyze metatranscriptomes.
All parts of this workflow are housed in their own repositories and imported via WDL v1.0 https importing. The following repositories are used in this workflow:
0.0.6
To run this workflow you will need a Docker (Docker ≥ v2.1.0.3) instance and cromwell. All the third party tools are pulled from Dockerhub.
cromwell ≥ 54
bbtools ≥ v38.94
Python ≥ v3.7.12
pandas ≥ v1.0.5 (python package)
gffutils ≥ v0.10.1 (python package)
metaT uses the same database uses for metagenome annotation. See README here for required databases. For QC databases see here
The submit script will request a node and launch the Cromwell. The Cromwell manages the workflow by using Shifter to run applications.
java -Dconfig.file=wdls/shifter.conf -jar /full/path/to/cromwell-XX.jar run -i input.json /full/path/to/wdls/metaT.wdl
microbiomedata/meta_t:0.0.5bryce911/bbtools:38.86
{
"metaT.input_files": ["./test_data/small_test/test_small_interleave.fastq.gz"],
"metaT.project_id":"nmdc:xxxxxxx",
"metaT.strand_type": "aRNA"
}project_id: A unique name for your project or sample.input_files: Full path to the fastq file. The file must be intereleaved paired end fastq.strand_type: (optional) RNA strandedness, either left blank,aRNA, ornon_stranded_RNA
All outputs can be found in the outdir folder. There are following subfolders:
outdir/annotation: contains gff files from annotation run.outdir/assembly: contains FASTA files from assembly and BAM files where reads were mapped back to the contigs.outdir/readMapping: JSON files for sense and antisense that have records for feature, their annotations, read counts, ans associated statistics.outdir/readsQC: contains cleaned reads and a file with associated statistics.
The output file is a JSON formatted file called out.json with JSON records that contains reads and information from annotation. An example JSON record:
{
"featuretype": "CDS",
"seqid": "nmdc:xxxxxxx_001",
"id": "nmdc:xxxxxxx_001_1_588",
"source": "Prodigal v2.6.3_patched",
"start": 1,
"end": 588,
"length": 588,
"strand": "+",
"frame": "0",
"product": "hypothetical protein",
"product_source": "Hypo-rule applied",
"sense_read_count": 25,
"mean": 5.0,
"median": 3.0,
"stdev": 6.1,
"antisense_read_count": 28,
"meanA": 7.14,
"medianA": 7,
"stdevA": 5.7
}
To test the workflow, we have provided a small test dataset and a step by step guidance. See test_data folder.