If you use this tool, please cite:
Arikan M, Muth T. (2024) gNOMO2: a comprehensive and modular pipeline for integrated multi-omics analyses of microbiomes. GigaScience, 13, giae038, https://doi.org/10.1093/gigascience/giae038
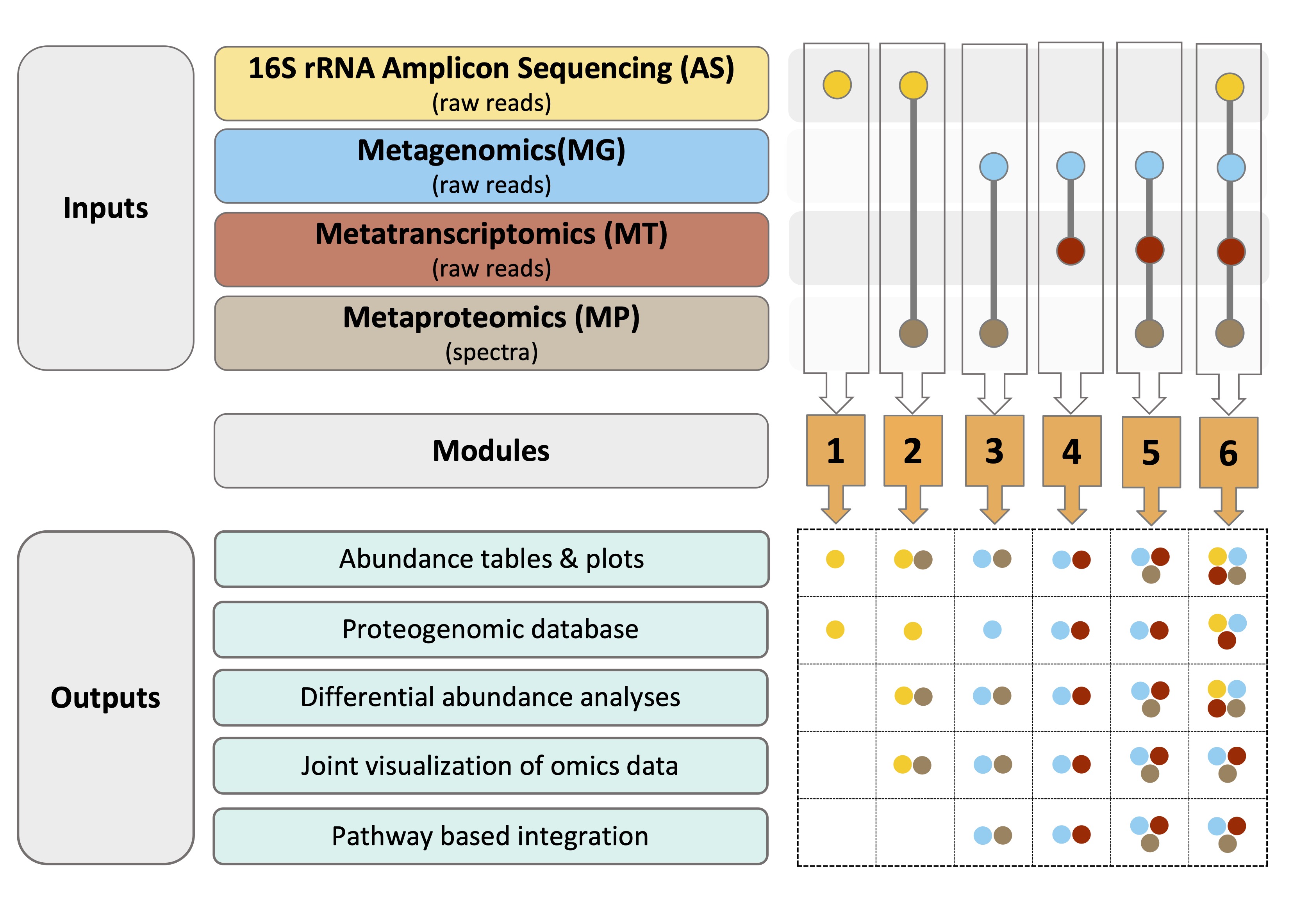
gNOMO2 comprises six modules, each tailored for specific omics data combination (shown below). Module 1 accepts 16S rRNA gene amplicon sequencing (AS) data as input and generates a protein database suitable for metaproteomics studies, a taxa abundance plot and a phyloseq object that can be used for downstream analysis in other microbiome tools. Modules 2 to 6 handle different combinations of AS, metagenomics (MG), metatranscriptomics (MT), and metaproteomics(MP) data, creating omics-specific protein databases, abundance tables, plots, differential abundance analysis results, joint visualization and pathway-level integration analysis results.
To use gNOMO2, ensure you have conda and snakemake installed:
1. Install conda: If you do not have conda installed, install conda.
2. Create a Snakemake environment in conda:
conda create -n snakemake bioconda::snakemake=7.15.2 conda-forge::mamba
3. Clone gNOMO2 repository: If you do not have git installed, install git.
git clone --recursive https://github.com/muzafferarikan/gNOMO2.git
Note: Once conda and snakemake are set up, gNOMO2 manages the installation of all other tools and dependencies automatically in their respective environments during the first run.
Copy your raw data to the relevant subfolders within the data directory:
- If you have amplicon sequencing data, copy your files to
data/AS/raw - If you have metagenomics data, copy your files to
data/MG/raw - If you have metatranscriptomics data, copy your files to
data/MT/raw - If you have metaproteomics data, copy your files to
data/MP/spectra.
Important: Please check sample format requirements below:
| Data | Library Layout | Sample Name Format |
|---|---|---|
| AS | PE SE |
samplename_1.fastq.gz, samplename_2.fastq.gz samplename_1.fastq.gz |
| MG | PE | samplename_1.fastq.gz, samplename_2.fastq.gz |
| MT | PE SE |
samplename_1.fastq.gz, samplename_2.fastq.gz samplename_1.fastq.gz |
| MP | DDA | samplename.mgf |
gNOMO2 requires a metadata file to perform sample group comparisons. Create a tab delimited metadata file (name it metadata.txt) containig information about samples and copy it to the resources folder.
Important:
- The name of the first column in metadata file must be "SampleID"
After copying your data and metadata, run the following script from your main gNOMO2 project folder to generate a config file:
bash workflow/scripts/prepare_config.sh
This script generates a config.yaml file within config folder based on contents of data directory. Review and modify analysis parameters in this file if you need.
Once setup is complete, follow these steps to run gNOMO2:
1. Activate your snakemake environment in conda:
conda activate snakemake
2. Run gNOMO2:
Execute the following command from your project folder:
snakemake -s workflow/Snakefile --cores 2 --use-conda
Note: Adjust the --cores value to reflect the number of cores available.
To run gNOMO2 on a cluster:
1. Configure the cluster settings:
Edit the provided gnomo2_slurm_template.sh file in the main gNOMO2 folder according to your cluster settings.
2. Run gNOMO2:
Execute the following command from your home directory in the cluster environemnt:
sbatch path/to/gNOMO2/gnomo_slurm_template.sh
When gNOMO2 pipeline starts, it generates a results folder within your project directory, containing both final and intermediate outputs.
The final folder includes:
-
Integrated multi-omics analysis results (
integrated)- Differential abundance analysis results for each omics dataset (
diff_abun) - Joint-visualization results (
combi) - Pathway level integration results (
pathview) - A proteogenomic database (
prot_db)
- Differential abundance analysis results for each omics dataset (
-
Results for each omics dataset within folders named accordingly (
AS,MG,MT,MP).- Abundance tables
- Taxonomy tables
- Phyloseq objects
- Abundance plots
These files are suitable for further analyses using other microbiome analysis tools.
intermediate folder contains outputs of each step executed by the gNOMO2 pipeline.