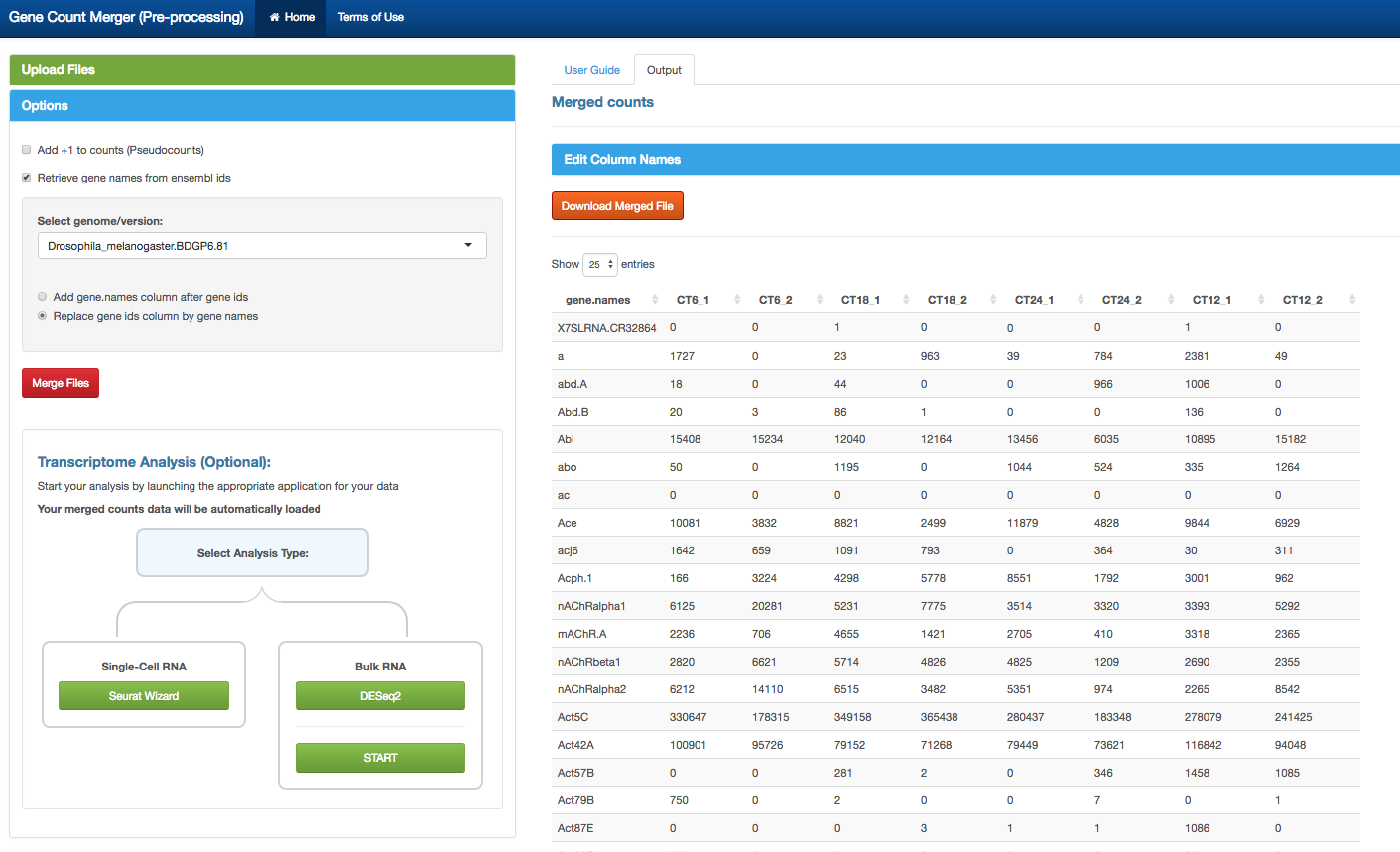
- This is a simple preprocessing tool to merge individual gene count files (Eg. output count files from htseq)
- This tool is a part of the NASQAR toolbox
- Pre-print: NASQAR: A web-based platform for High-throughput sequencing data analysis and visualization
NOTE: first column must contain the genes. If the gene columns do not match in all files, this tool will not work
You can try it online at http://nasqar.abudhabi.nyu.edu/GeneCountMerger
- Merge individual sample count files. See Sample Input Files below for more details
- Or merge multiple matrices
- Convert ensembl gene IDs to gene names
- Option to choose from available genome/versions
- If genome/version is not available in the options and you have a .gtf file for your genome follow these instructions.
- Option to add pseudocounts (+1)
- Download merged counts file in .csv format
- Transcriptome Analysis (Optional) after merging your counts:
- Use our Seurat Wizard to carry out single-cell RNA analysis
- Use DESeq2 or START apps to carry out bulk RNA analysis