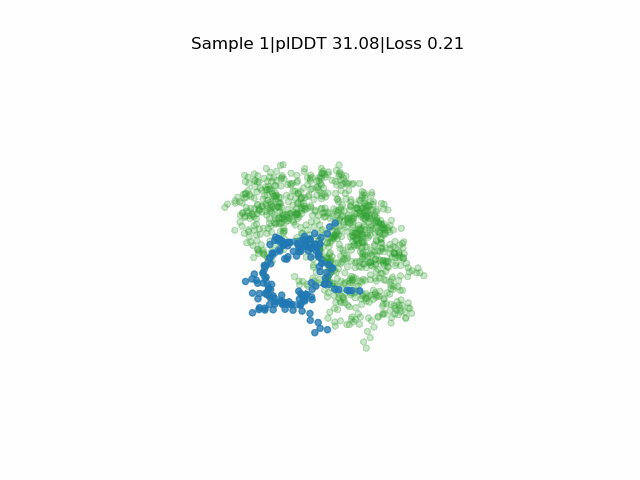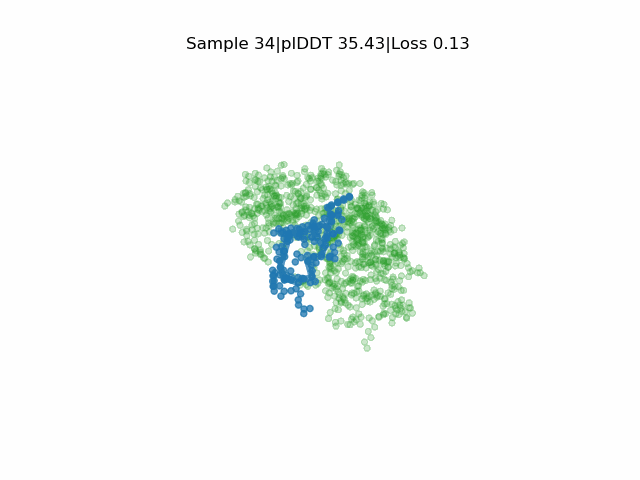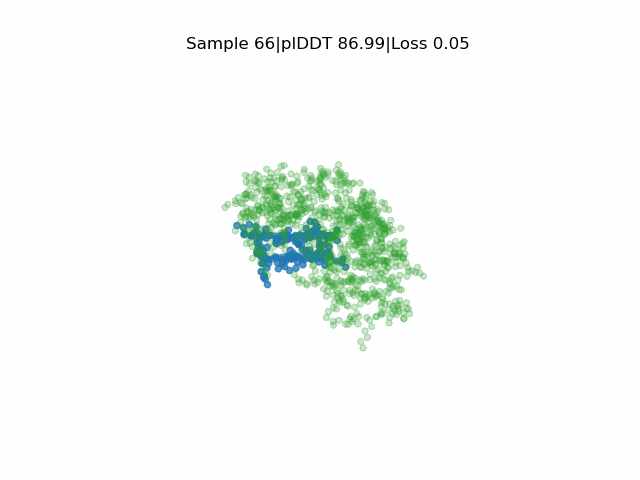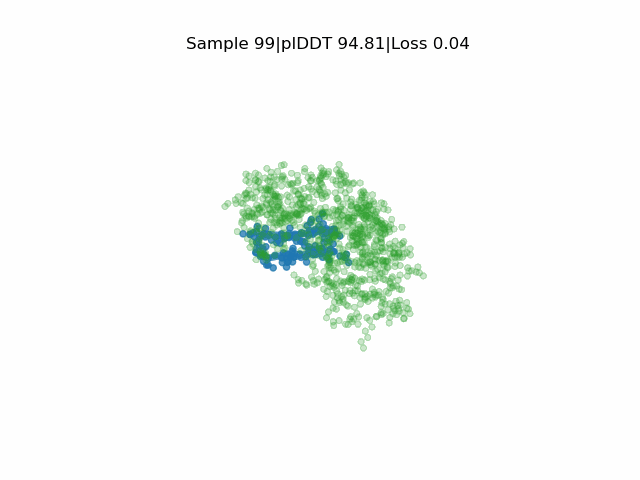
In silico directed evolution of peptide binders
Read more here
EvoBind (v2) designs novel peptide binders based only on a protein target sequence. It is not necessary to specify any target residues within the protein sequence or the length of the binder (although this is possible). Cyclic binder design is also possible.
Linear binder success rate = 46%
Cyclic binder success rate = 75%
[Success rate = probability of obtaining a binder with Kd≤μM affinity (according to SPR analysis) from a single sequence selection.]
WT affinity = 35 nM
Best linear (from 13) = 19 nM
Best cyclic (from 4) = 0.26 nM
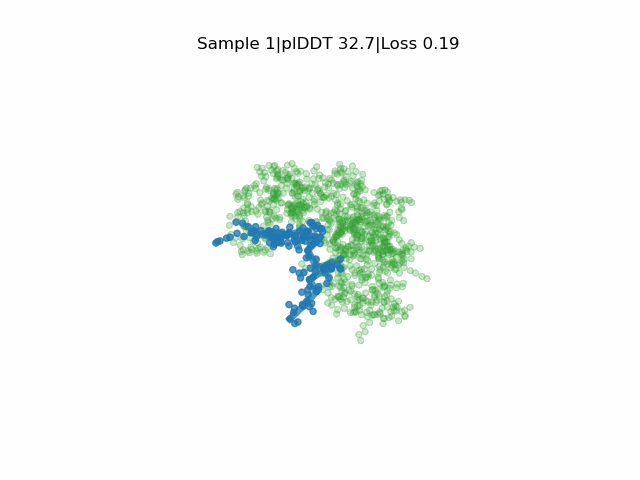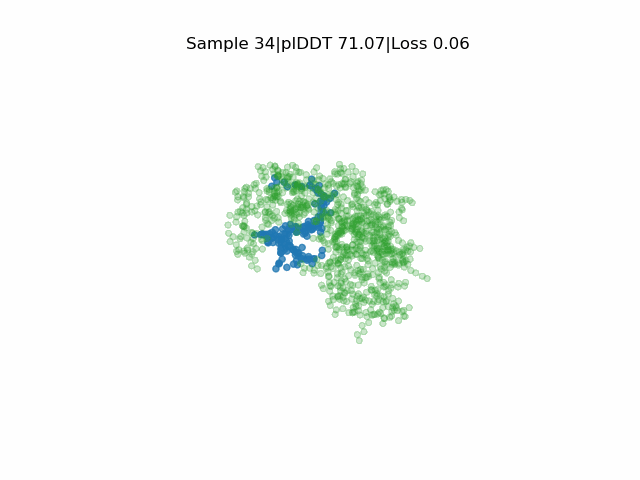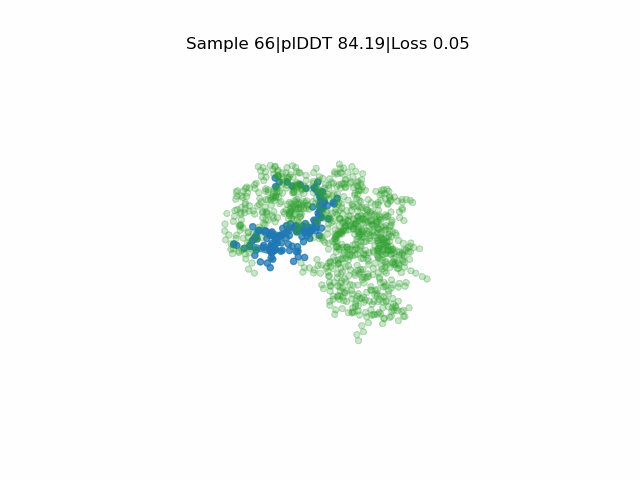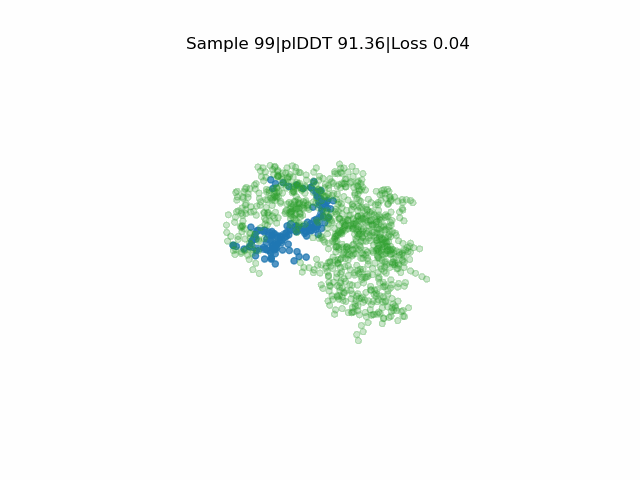
EvoBind2 accounts for adaptation of the receptor interface structure to the peptide being designed during optimisation. This consideration of flexibility is crucial for binding. EvoBind is the first protocol that only relies on a protein sequence to design a binder and the only one with experimentally verified cyclic design capacity.
Receptor in green and peptide in blue.
EvoBind2 is based on AlphaFold2, which is available under the Apache License, Version 2.0 and so is EvoBind2, which is a derivative thereof.
The AlphaFold2 parameters are made available under the terms of the CC BY 4.0 license and have not been modified.
You may not use these files except in compliance with the licenses.
It is possible to run EvoBind2 online in the Google colab here
Before beginning the process of setting up this pipeline on your local system, make sure you have adequate computational resources. Make sure you have an available GPU as this will speed up the prediction process substantially compared to using a CPU. EvoBind2 assumes you have NVIDIA GPUs on your system, readily available. A Linux-based system is assumed.
To setup this pipeline, clone this github repository:
git clone https://github.com/patrickbryant1/EvoBind.git
Then do
bash setup.sh
This script fetches the AlphaFold2 parameters, installs a conda env and downloads uniclust30_2018_08 which is used to generate the receptor MSA.
To design binders the following needs to be specified:
Receptor fasta sequence
Optional arguments:
Peptide length - default=10
Target residues within the raceptor sequence - default=all
If you want to design a cyclic peptide, add the flag --cyclic_offset=1 in the design script when calling mc_design.py. Based on cyclic offset.
A test case is provided in design_local.sh.
This script can be run by simply doing:
bash design_local.sh
See src/AFM_eval for instructions
If you use EvoBind in your research, please cite
- Li Q, Vlachos E.N., Bryant P. Design of linear and cyclic peptide binders of different lengths from protein sequence information. bioRxiv. 2024. p. 2024.06.20.599739. doi:10.1101/2024.06.20.599739
- Bryant P, Elofsson A. EvoBind: in silico directed evolution of peptide binders with AlphaFold. bioRxiv. 2022. p. 2022.07.23.501214. doi:10.1101/2022.07.23.501214
EvoBind (v1) is available here