===========================================================================================
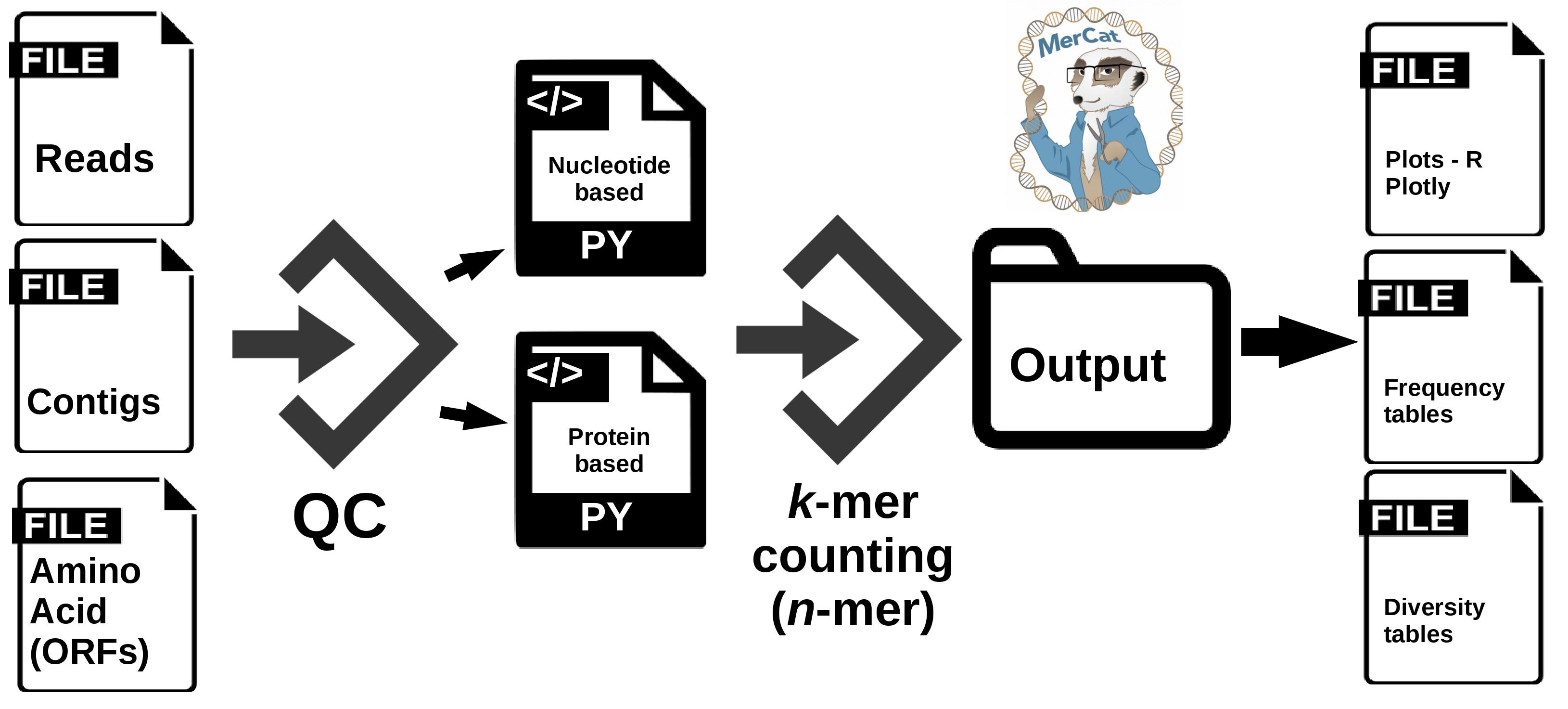
MerCat: python code for versatile k-mer counter and diversity estimator for database independent property analysis (DIPA) obtained from metagenomic and/or metatranscriptomic sequencing data
Installing MerCat:
- Available via Anaconda: Enable BioConda repo and run
conda install mercat - We do not have a pip installer available as of now. If you would like to use pip, please install the
modules listed in
dependencies.txtvia pip and runpython setup.py installfor setting up mercat.
- -i I path-to-input-file
- -f F path-to-folder-containing-input-files
- -k K kmer length
- -n N no of cores [default = all]
- -c C minimum kmer count [default = 10]
- -pro run mercat on protein input file specified as .faa
- -q tell mercat that input file provided are raw nucleotide reads as [.fq, .fastq]
- -p run prodigal on nucleotide assembled contigs. Must be one of ['.fa', '.fna', '.ffn', '.fasta']
- -t [T] Trimmomatic options
- -s Data split size for large files (default is 100 Mb file size)
- -h, --help show this help message
By default mercat assumes that inputs provided is one of ['.fa', '.fna', '.ffn', '.fasta']
Example: To compute all 3-mers, run
mercat -i test.fa -k 3 -n 8 -c 10 -p
The above command:
-
Runs prodigal on
test.fa, then runs mercat on the resulting protein file. -
Results are generally stored in input-file-name_{protein|nucleotide}.csv and input-file-name_{protein|nucleotide}_summary.csv
test_protein.csvandtest_protein_summary.csvin this example
-
test_protein_summary.csvcontains kmer frequency count, pI, Molecular Weight, and Hydrophobicity metrics for all unique kmers across all sequences intest.fa -
test_protein_diversity_metrics.txtcontaining the alpha diversity metrics. -
test_protein.csvcontains kmer frequency count, pI, Molecular Weight, and Hydrophobicity metrics for individual sequences.NOTE: We disabled the code that generates this file since computing k-mer counts for individual sequences was getting very expensive in terms of time & memory usage for large input files.
-
mercat -i test.fq -k 3 -n 8 -c 10 -q
Runs mercat on raw nucleotide read (.fq or .fastq) -
mercat -i test.fq -k 3 -n 8 -c 10 -q -t
Runs trimmomatic on raw nucleotide reads (.fq or .fastq), then runs mercat on the trimmed nucleotides -
mercat -i test.fq -k 3 -n 8 -c 10 -q -t 20
Same as above but can provide the quality option to trimmomatic -
mercat -i test.fq -k 3 -n 8 -c 10 -q -t 20 -pRun trimmomatic on raw nucleotide reads, then run prodigal on the trimmed read to produce a protein file which is then processed by mercat -
mercat -i test.fna -k 3 -n 8 -c 10
Run mercat on nucleotide input - one of ['.fa', '.fna', '.ffn', '.fasta'] -
mercat -i test.fna -k 3 -n 8 -c 10 -p
Run prodigal on nucleotide input, generate a .faa protein file and run mercat on it -
mercat -i test.faa -k 3 -n 8 -c 10 -pro
Run mercat on a protein input (.faa) -
All the above examples can also be used with
-f input-folderinstead of-i input-fileoption- Example:
mercat -f /path/to/input-folder -k 3 -n 8 -c 10--- Runs mercat on all inputs in the folder
- Example:
-
To save working memory (RAM) on low RAM computers or >2 GB files use '-s option' to split/chunk the file
- Example:
mercat -i test.fna -k 3 -n 8 -c 10 -s 50--Runs mercat in nucleotide mode splitting file into 50 MB pieces
- Example:
If you are publishing results obtained using MerCat, please cite:
White III RA, Panyala A, Glass K, Colby S, Glaesemann KR, Jansson C, Jansson JK. (2017) MerCat: a versatile k-mer counter and diversity estimator for database-independent property analysis obtained from metagenomic and/or metatranscriptomic sequencing data. PeerJ Preprints 5:e2825v1 https://doi.org/10.7287/peerj.preprints.2825v1
Please send all queries to Richard Allen White III <rwhit101@uncc.edu> or <raw937@gmail.com>