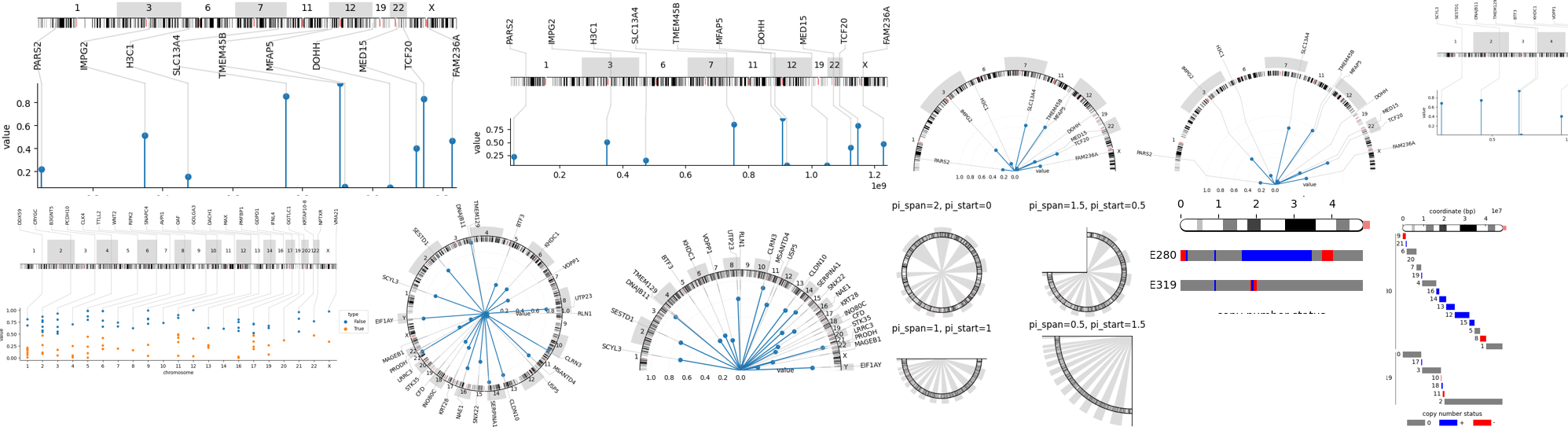
🏷️Annotations on chromosomes
🧬Visualization of genome, set of chromosomes, a chromosome and an arm
🔥🗺️Visualization of heatmaps along chromosome arm
🔌Integration of custom plots e.g. made using seaborn
📈🍭Integration of basic plots e.g. stem/lollipop plot
pip install chrov # with basic dependencies
With additional dependencies as required:
pip install chrov[dev] # for local testing
- Using BibTeX:
@software{Dandage_chrov,
title = {chrov: Chromosome Visualization library in python},
author = {Dandage, Rohan},
year = {2023},
url = {https://zenodo.org/doi/10.5281/zenodo.10211265},
version = {v0.0.1},
note = {The URL is a DOI link to the permanent archive of the software.},
}
-
Using citation information from CITATION.CFF file.
- Set subplot sizes by default: automate setting off and offy.
- Compatibility of seaborn plots with polar layout.
- Layering the interval vizualizations on the polar layout.
- Connection styles.
- Other features and improvements.
Annotations.
annot_labels(
ax_chrom: Axes,
data: DataFrame,
colx: str,
chrom_y: float,
col_label: str,
loc: str = 'out',
col_start: str = None,
ax: Axes = None,
coly: str = None,
col_labelx: str = 'label x',
color: str = 'darkgray',
yoff_scales: float = None,
off_labels_segments: float = 20,
scale_polar: float = 1.5,
fig: Figure = None,
test: bool = False
) → AxesAnnot labels e.g. gene names
Args:
ax_chrom(plt.Axes): subplot with the chromosome plotcol_label(str): column with the labelsloc(str, optional): locations. Defaults to 'out'.color(str, optional): color. Defaults to 'darkgray'.yoff_scales(float, optional): y offset. Defaults to None.off_labels_segments(float, optional): offset for the label segments. Defaults to 20.scale_polar(float, optional): scale for the polar plot. Defaults to 1.5.fig(plt.Figure, optional): figure. Defaults to None.test(bool, optional): test-mode. Defaults to False.
Returns:
plt.Axes: subplot
show_segments(
ax: Axes,
y: float,
offy: float,
kind: str = 'arrows',
segments: dict = None,
segments_kws: dict = {},
offytext: float = 0.2,
arrow_kws: dict = {'color': 'k', 'lw': 1, 'alpha': 1, 'arrowstyle': '<->'},
test: bool = False,
**kws_annotate
) → AxesShow segments aligned to chromosome arm.
Args:
ax(plt.Axes): subplotdata(pd.DataFrame): input datasize(int): size of the segments
Chromosome plots
to_polar(
a: list,
range1: list = None,
range2: list = None,
interval: int = None
) → listTo polar coordinates
Args:
a(list): Coordinatesrange1(list, optional): range1. Defaults to None.range2(list, optional): range2. Defaults to None.interval(int, optional): interval size. Defaults to None.
Raises:
ValueError: Coordinates format
Returns:
list: rescaled coordinates
plot_arm(
data: DataFrame,
arc: bool = False,
col_start: str = 'start',
col_end: str = 'end',
y: float = 0,
lw: float = 10,
ec: str = 'k',
pi_span: float = 1,
pi_start: int = 0,
pi_end: int = None,
polar_smoothness_scale: float = 1,
kws_pre_xys: dict = None,
figsize: list = None,
ax: Axes = None,
test: bool = False,
solid_capstyle='round'
) → AxesPlot chromosome arm.
Args:
data(pd.DataFrame): input table.y(float, optional): y position. Defaults to 0.lw(float, optional): line width. Defaults to 20.ec(str, optional): edge color. Defaults to 'k'.ax(plt.Axes, optional): subplot. Defaults to None.test(bool, optional): test-mode. Defaults to False.
Returns:
plt.Axes: subplot
plot_chrom(
data: DataFrame,
arc: bool = False,
col_start: str = 'start',
col_end: str = 'end',
col_arm: str = 'arm',
pi_span: float = 1,
pi_start: int = 0,
pi_end: int = None,
ax: Axes = None,
figsize: list = None,
**kws_plot_arm
) → AxesPlot a chromosome
Args:
data(pd.DataFrame): cytobandsarc(bool, optional): arc/polar mode. Defaults to False.col_start(str, optional): column with start positions. Defaults to 'start'.col_end(str, optional): column with end positions. Defaults to 'end'.col_arm(str, optional): column with arm names. Defaults to 'arm'.pi_span(float, optional): pi span. Defaults to 1.pi_start(int, optional): pi start. Defaults to 0.pi_end(int, optional): pi end. Defaults to None.ax(plt.Axes, optional): subplot. Defaults to None.figsize(list, optional): sigure size. Defaults to None.
Returns:
plt.Axes: subplot
plot_chroms(
data: DataFrame,
arc=True,
chromosomes: list = None,
col_start='start',
col_end='end',
col_arm='arm',
span_color: str = '#dcdcdc',
span_color_alpha_scale: float = 1,
pi_span: float = 1,
pi_start: int = 0,
pi_end: int = None,
show_labels: bool = True,
show_vline: bool = True,
label_y: str = None,
test: bool = False,
ax: Axes = None,
figsize: list = None,
out_data: bool = False,
**kws_plot_arm
)Plot chromosomes joined.
Args:
data(pd.DataFrame): cytonbandsarc(bool, optional): arc/polar mode. Defaults to True.chromosomes(list, optional): chromosomes. Defaults to None.col_start(str, optional): column with start position. Defaults to 'start'.col_end(str, optional): column with end position. Defaults to 'end'.col_arm(str, optional): column with chromosome arm names. Defaults to 'arm'.span_color(str, optional): span color. Defaults to 'whitesmoke'.span_color_alpha_scale(float, optional): span color transparency scale. Defaults to 1.pi_span(float, optional): pi span. Defaults to 1.pi_start(int, optional): pi start angle. Defaults to 0.pi_end(int, optional): pi end angle. Defaults to None.show_labels(bool, optional): show labels. Defaults to True.show_vline(bool, optional): show vertical line. Defaults to True.label_y(str, optional): label y. Defaults to None.test(bool, optional): test-mode. Defaults to False.ax(plt.Axes, optional): subplot. Defaults to None.figsize(list, optional): figure size. Defaults to None.out_data(bool, optional): output data. Defaults to False.
annot_chroms(
data: DataFrame,
chromosomes: list,
ax_chrom: Axes = None,
chrom_y: float = 0,
kws_add_ax: dict = {},
test: bool = False,
**kws_plot
) → AxesAdd a subplot with the chromosome.
Args:
data(pd.DataFrame): table with cytobandschromosomes(list): chromosomesax_chrom(plt.Axes, optional): subplot with chromosome plot. Defaults to None.chrom_y(float, optional): chromosome y-position. Defaults to 0.kws_add_ax(dict, optional): keyword parameters provided to_add_ax. Defaults to {}.test(bool, optional): test mode. Defaults to False.
Returns:
plt.Axes: subplot
plot_with_chroms(
data: DataFrame,
cytobands: DataFrame,
kind: str,
colx: str,
coly: str,
col_label: str,
va: str,
col_start: str = None,
xkind: str = 'loci',
coffy: str = None,
off: float = None,
offy: float = None,
chrom_y: float = 0,
arc: bool = True,
pi_span: float = 1,
pi_start: int = 0,
pi_end: int = None,
fig: Figure = None,
figsize: list = None,
ax_data: Axes = None,
kws_seaborn: dict = {},
kws_annot_chroms: dict = {},
kws_annot_labels: dict = {},
test: bool = False
) → FigurePlot with chromosomes.
Args:
data(pd.DataFrame): input tablecytobands(pd.DataFrame): cytobandskind(str): kind of plotcolx(str): column with x valuescoly(str): column with y valuescol_label(str): column with labelsva(str): vertical alignmentcol_start(str, optional): column with start positions. Defaults to None.xkind(str, optional): kind of x values. Defaults to 'loci'.off(float, optional): offset scale of the chromosome plot. Defaults to None.offy(float, optional): offset y of the chromosome plot. Defaults to None.chrom_y(float, optional): chromosome y-position. Defaults to 0.arc(bool, optional): arc/polar plot or linear/rectangular one. Defaults to True.pi_span(float, optional): pi span. Defaults to 1.pi_start(int, optional): pi start. Defaults to 0.pi_end(int, optional): pi end. Defaults to None.fig(plt.Figure, optional): figure. Defaults to None.figsize(list, optional): figure size. Defaults to None.ax_data(plt.Axes, optional): subplot with the data plot. Defaults to None.kws_seaborn(dict, optional): keyword parameters to seaborn plot. Defaults to {}.kws_annot_chroms(dict, optional): keyword parameters to the chromosome plot. Defaults to {}.kws_annot_labels(dict, optional): keyword parameters to the annotations of the labels. Defaults to {}.test(bool, optional): test mode. Defaults to False.
Returns:
plt.Figure: figure
plot_seaborn(
data: DataFrame,
kind: str,
colx: str,
coly: str,
range1_chroms: list,
arc: bool = True,
pi_span: float = 1,
pi_start: int = 0,
pi_end: int = None,
figsize: list = None,
ax: Axes = None,
fig: Figure = None,
**kws_plot
) → tupleplot_seaborn summary
Args:
data(pd.DataFrame): input datakind(str): kind of plot, seaborn function namecoly(str): column with y valuesrange1_chroms(list): input range of chromosomesarc(bool, optional): arc/polar or linear/rectangular plots. Defaults to True.pi_span(float, optional): pi span. Defaults to 1.pi_start(int, optional): pi start position. Defaults to 0.pi_end(int, optional): pi end position. Defaults to None.figsize(list, optional): figure size. Defaults to None.ax(plt.Axes, optional): subplot. Defaults to None.fig(plt.Figure, optional): figure. Defaults to None.
Returns:
tuple: subplot and data
TODOs: 1. set rlabel position.
heatmaps_strips(
data: DataFrame,
strips_kws: dict,
fig: Figure = None,
axs: list = None,
kws_subplots: list = {}
) → tuplePlot heatmap strips
Args:
data(pd.DataFrame): input datastrips_kws(dict): keyword arguments provided to stripsfig(plt.Figure, optional): figure. Defaults to None.axs(list, optional): subplots. Defaults to None.kws_subplots(list, optional): keyword arguments provided to subplots. Defaults to {}.
Returns:
tuple: figure and subplots
plot_ranges(
data: DataFrame,
col_id: str,
col_start: str,
col_end: str,
end: int,
start: int = 0,
hue: str = None,
y=None,
kind=None,
cytobands: dict = None,
cytobands_y: float = None,
col_groupby: str = None,
col_label: str = None,
colors: dict = None,
lw: int = 10,
zorders: dict = None,
show_segments: bool = False,
xtick_interval: float = None,
test: bool = False,
ax: Axes = None
) → AxesPlot ranges.
Args:
data(pd.DataFrame): input data.col_id(str): column with ids.col_start(str): column with start co-ordinates.col_end(str): column with end co-ordinates.end(int): end position for the plotstart(int, optional): start position for the plot. Defaults to 0.hue(str, optional): column with color. Defaults to None.y(type, optional): column with y positions. Defaults to None.cytobands(dict, optional): cytobands to plot the chromosomes. Defaults to None.cytobands_y(float, optional): cytobands y-position. Defaults to None.col_groupby(str, optional): column to group by. Defaults to None.col_label(str, optional): column with labels. Defaults to None.colors(dict, optional): colors. Defaults to None.lw(int, optional): line width. Defaults to 10.zorders(dict, optional): z-orders. Defaults to None.show_segments(bool, optional): show segments. Defaults to False.xtick_interval(float, optional): x tick intervals. Defaults to None.test(bool, optional): test-mode. Defaults to False.ax(plt.Axes, optional): subplot. Defaults to None.
Raises:
ValueError: if kind is not 'split','separate' or 'joined'
Returns:
plt.Axes: subplot