CHOLAR short for CHaracterization Of LncRNA from rAw Reads).
It extracts the list of known and novel lncRNAs from any RNA-seq dataset. As of now, the tool is resticted to human datasets, but we are working hard to include other model organism too.
brew is a pre-requistic. For information on brew visit https://brew.sh/
Download the zip file for CHOLAR from here or
Download the tar file for CHOLAR from here
After download unzip the file using:
unzip Download the zip file for CHOLAR from here or
Download the tar file for CHOLAR from here
After download unzip the file using:
unzipExecute the configure file on LINUX/UNIX system using terminal:
bash configure.shThis will prompt to give password. Enter it and let the script run.
Execute CHOLOAR GUI file using:
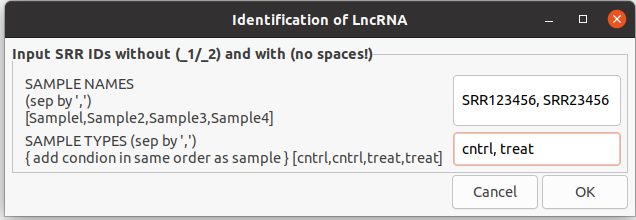
bash CHOLAR_GUI.sh1.1 This will pop up a window to input sample names in the box and sample condition separated by comma.
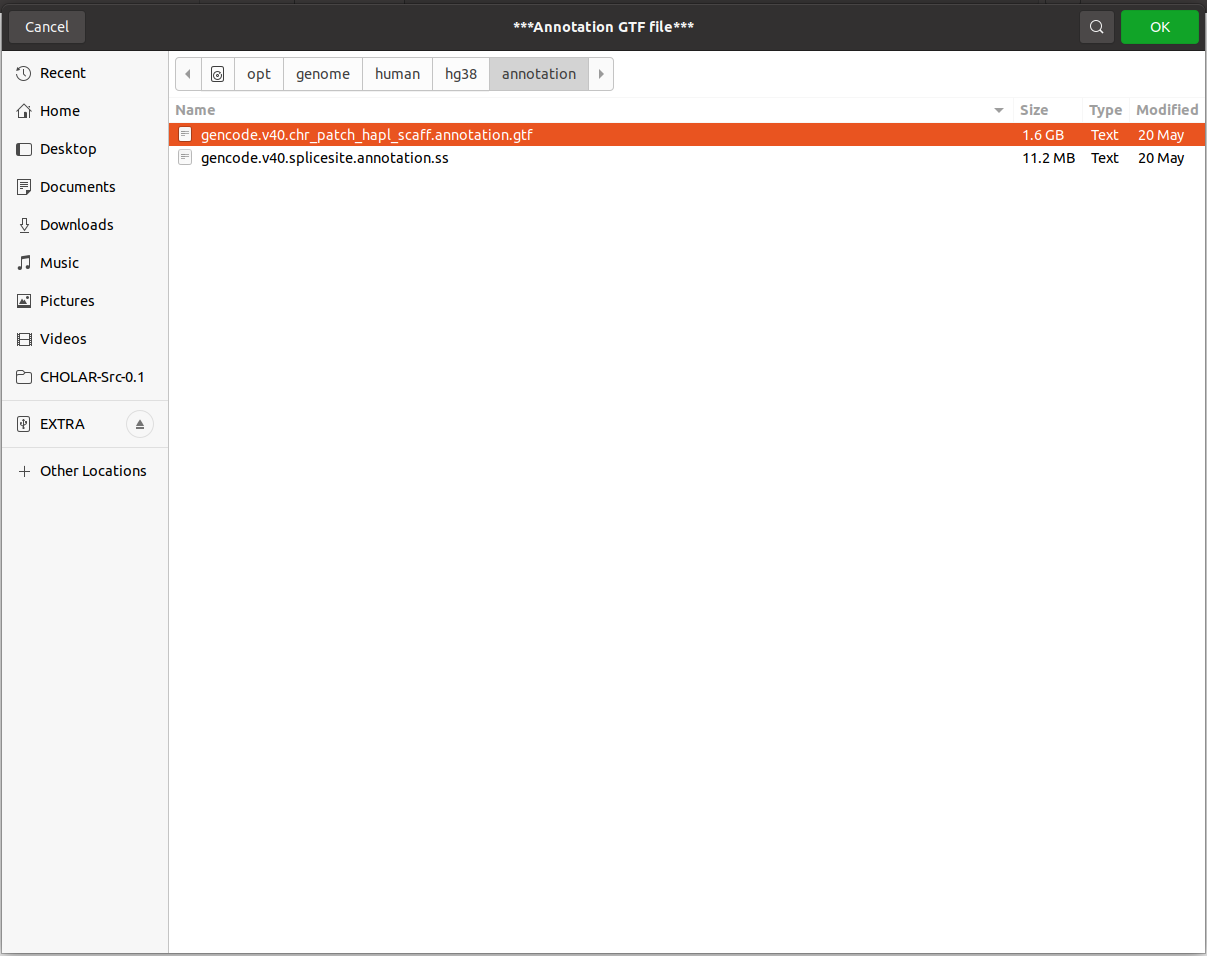
2.2 Then the Annotation file selection will appear (if it appears daunting, just press ok in both dialog box).
CHOLAR is developed by Haneesh J, Anubha Dey and Manjari Kiran (Department of Systems and Computational Biology, School of Life Sciences, University of Hyderabad, India).
If you use CHOLAR in your publications, you can cite the package as follows:
CHOLAR: Characterization of LncRNA from raw reads.
or in BiBTex:
See the LICENSE file for license rights and limitations.
This work is supported by SERB Startup Grant (SRG/2020/002146) from Department of Science and Technology (DST), India.