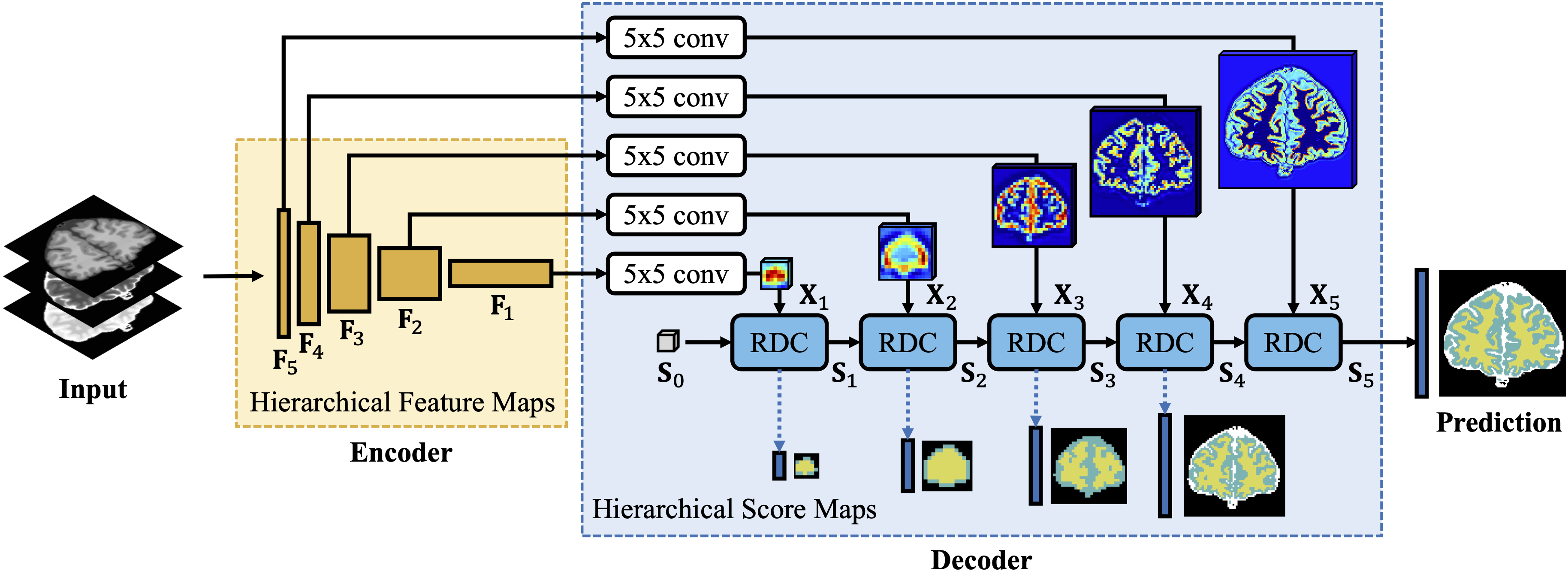
This is the PyTorch implementation for AAAI 2020 paper Segmenting Medical MRI via Recurrent Decoding Cell by Ying Wen, Kai Xie, Lianghua He.
Recurrent Decoding Cell (RDC) is a novel feature fusion unit used in the encoder-decoder segmentation network for MRI segmentation. RDC leverages convolutional RNNs (e.g. ConvLSTM, ConvGRU) to memorize the long-term context information from the previous layers in the decoding phase. The RDC based encoder-decoder network named Convolutional Recurrent Decoding Network (CRDN) achieves promising semgmentation reuslts -- 99.34% dice score on BrainWeb, 91.26% dice score on MRBrainS, and 88.13% dice score on HVSMR. The model is also robust to image noise and intensity non-uniformity in medical MRI.
- FCN
- SegNet
- UNet
- CRDN (Ours) with different encoders
- U-Net(decoder) with VGG16(encoder) (VGGUNet)
- U-Net(decoder) with ResNet50(encoder) (ResNet50UNet)
- FCN(decoder) with U-Net-backbone(encoder) (UNetFCN)
- FCN(decoder) with ResNet50(encoder) (ResNet50FCN)
- SegNet(decoder) with U-Net-backbone(encoder) (UNetSegNet)
- pytorch == 1.1.0
- torchvision == 0.2.2.post3
- matplotlib == 2.1.0
- numpy == 1.11.3
- tqdm == 4.31.1
One-line installation
pip install -r requirements.txt
Setup config
model:
arch: <name> [options: 'FCN, SegNet, UNet, VGG16RNN, ResNet50RNN, UNetRNN, VGGUNet, ResNet50UNet, UNetFCN, ResNet50FCN, UNetSegNet']
data:
dataset: <name> [options: 'BrainWeb, MRBrainS, HVSMR']
train_split: train
val_split: val
path: <path/to/data>
training:
gpu_idx: 0
train_iters: 30000
batch_size: 1
val_interval: 300
n_workers: 4
print_interval: 100
optimizer:
name: <optimizer_name> [options: 'sgd, adam, adamax, asgd, adadelta, adagrad, rmsprop']
lr: 6.0e-4
weight_decay: 0.0005
loss:
name: 'cross_entropy'
lr_schedule:
name: <schedule_type> [options: 'constant_lr, poly_lr, multi_step, cosine_annealing, exp_lr']
<scheduler_keyarg1>:<value>
# Resume from checkpoint
resume: <path_to_checkpoint>
# model save path
model_dir: <path_to_save_model>
testing:
# trained model path
trained_model: <path_to_trained_model>
# segmentation results save path
path: <path_to_results>
# if show boxplot results
boxplot: FalseTo train the model :
run train.py
To test the model :
run test.py
-
Some visualization results of the proposed CRDN and other encoding-decoding methods.
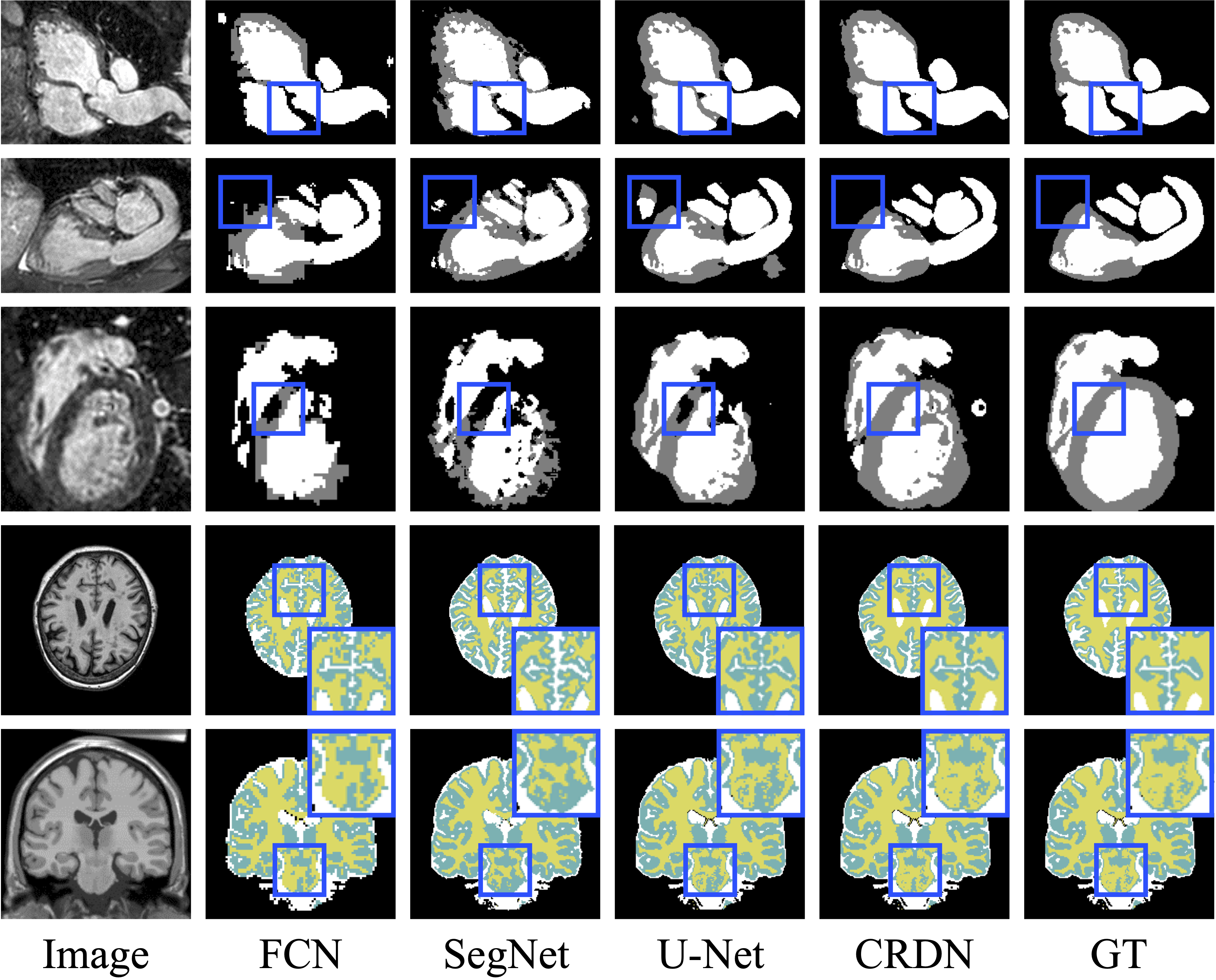
-
please refer to the paper for other experiments. (ablation study, comparisons, network robustness)
Special thanks for the github repository meetshah1995/pytorch-semseg for providing the semacntic segmentation algorithms in PyTorch.
Please cite these papers in your publications if it helps your research:
@inproceedings{wen2020segmenting,
title={Segmenting Medical MRI via Recurrent Decoding Cell.},
author={Wen, Ying and Xie, Kai and He, Lianghua},
booktitle={AAAI},
pages={12452--12459},
year={2020}
}For any problems, please contact kxie_shake@outlook.com