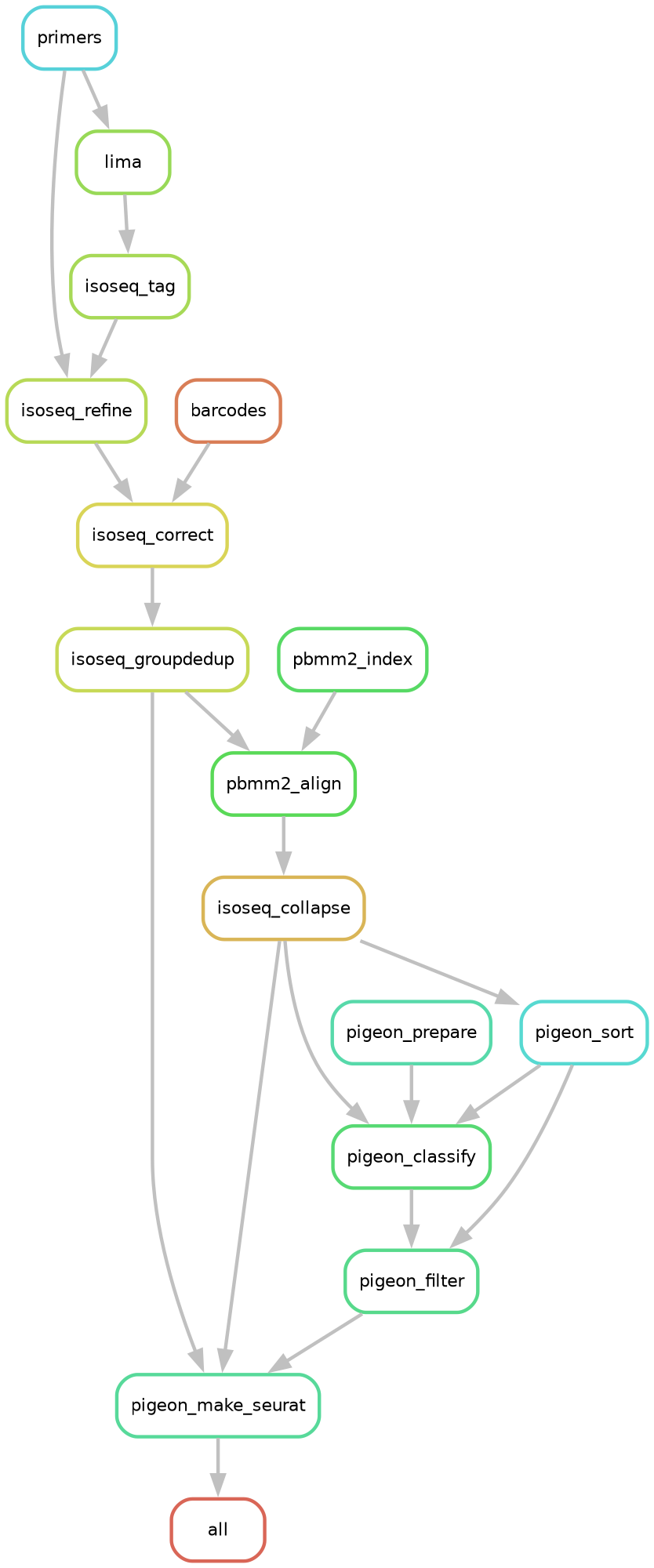
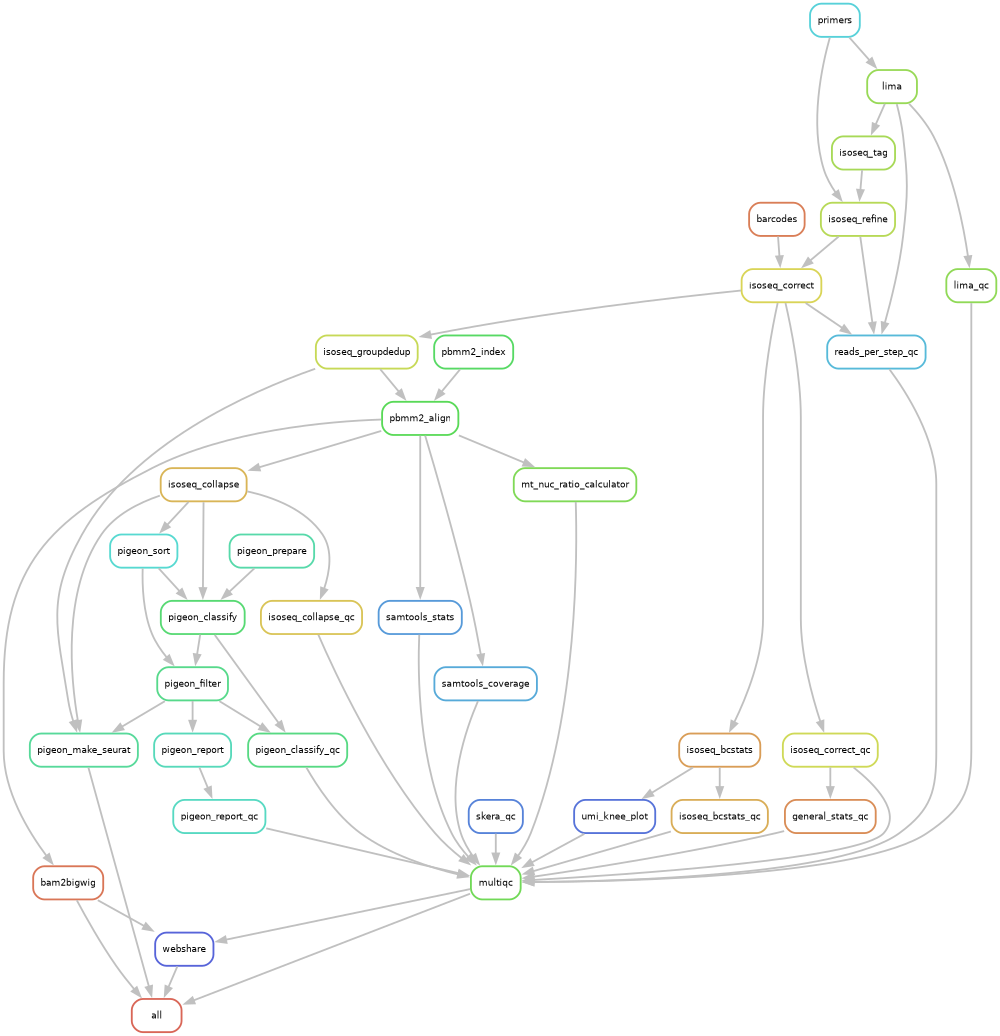
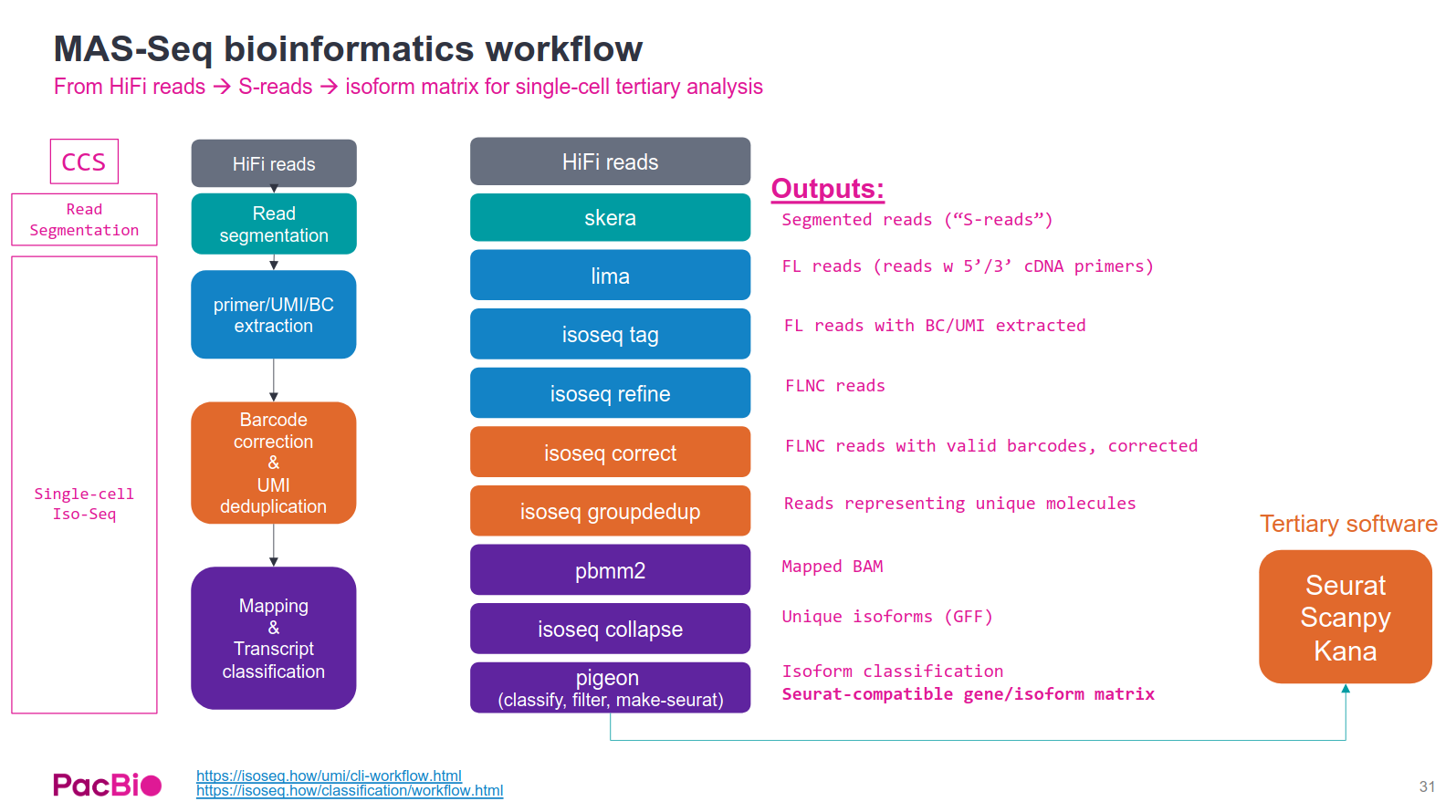
for PacBio single-cell MAS-seq data
Clone the repository:
git clone https://github.com/siebrenf/lore.git
Create the conda environment:
conda env create -n lore -f lore/requirements.yaml
conda activate lore
Install LoRE in the conda environment:
pip install -e ./lore
Change directory into the LoRE folder.
Activate the conda environment:
conda activate lore
Update the config.yaml.
- Adapters, primers and barcodes for the 5' Kinnex kit can be downloaded by LoRE, or can be placed inside the results directory.
- The results directory (as well as any other directory) can be set in the config.
The default results directory is
./results. - The genome and gene annotation need to be obtained manually.
You will need to specify their locations in the config, as well as the symbol for the mitochondria.
- If the
pigeon classifyoutput suggests the number of reads per cell is low, this may suggest the genome and/or gene annotation were insufficiently annotated.
- If the
- Additional documentation for most rules (steps) in the workflow can be found in the code.
- Optional outputs (currently) include bigwigs (for track visualization) and a QC report. Both are recommended, but adds (some) computational load.
Test your config:
snakemake --snakefile lore/Snakefile --configfile config.yaml --dry-run
Run your config:
nice snakemake --use-conda --snakefile lore/Snakefile --configfile config.yaml --resources parallel_downloads=1 mem_mb=100_000 -j 60 > log.txt 2>&1
- implement TE/RE detection using the output of either:
isoseq_groupdedup(an unaligned FASTA and BAM file)pbmm2_align(an aligned BAM file)- current settings:
- multimapped reads are included: reads are assigned to any number of locations (nice for TEs).
- unmapped reads are included.
- current settings:
- repositories of interest to this purpose have been marked below.
- learn more about the
pigeon classifyfilter settings (for the gene level, some filters may be more lenient). - integrate genomepy to get a genome & gene annotation.
- figure out the requirements for a "good" reference genome & gene annotation.
- de novo Repeat library construction pipelines
- https://doi.org/10.1186/s12864-021-08117-9
- https://github.com/kacst-bioinfo-lab/TE_ideintification_pipeline # nice workflow figure
- could be useful for non-model organisms/strains
- https://doi.org/10.1186/s12864-021-08117-9
- RE/TE pipelines
- SoloTE
- easy to use: use pbmm2 output
- https://doi.org/10.1038/s42003-022-04020-5
- https://github.com/bvaldebenitom/SoloTE
- TrEMOLO
- looks good + snakemake!
- https://doi.org/10.1186/s13059-023-02911-2
- https://github.com/DrosophilaGenomeEvolution/TrEMOLO
- tldr
- looks easy to use
- https://doi.org/10.1016/j.molcel.2020.10.024
- https://github.com/adamewing/tldr
- works with "partial detection, assembly and annotation of non-reference TE insertions"
- TELR
- looks good! pure python
- https://doi.org/10.1093/nar/gkac794
- https://github.com/bergmanlab/TELR
- "relies on the detection of abnormally mapped reads upon the reference genome that can be linked to a TE"
- teNanoporePipeline
- may be usable
- https://doi.org/10.1016/j.isci.2023.108214
- https://github.com/javiercguard/teNanoporePipeline
- Tools: RepeatMaster, Dfam
- LoRTE
- not usable (python 2!)
- https://doi.org/10.1186/s13100-017-0088-x
- dead link
- SoloTE