Our visualization tool multiSLIDE for hypothesis-driven, unbiased exploration of multi-omics data is described here:
Ghosh, S., Datta, A. & Choi, H. multiSLIDE is a web server for exploring connected elements of biological pathways in multi-omics data. Nat Commun 12, 2279 (2021). https://doi.org/10.1038/s41467-021-22650-x
multiSLIDE can be installed using the pre-built Docker image, the instructions are provided below.
The preferred way to run a local instance of multiSLIDE is using the pre-built Docker image available at Docker Hub, as follows:
-
Install Docker for Mac/Windows/Linux following the instructions here
-
Pull the multiSLIDE Docker image using the following command in Terminal(Mac) or Command Prompt(Windows) or the Linux Shell
$ docker pull soumitag/multislide:2.0
-
To launch multiSLIDE execute the following command:
$ docker run -d -p 8080:8080 soumitag/multislide:2.0
multiSLIDE should now be available at http://localhost:8080/multislide. Navigate to this link with your browser and start using multiSLIDE. Starting up the application might take a while, please take a sip of coffee and wait for a minute.
-
To stop multiSLIDE, first identify the name of the Docker container running multiSLIDE using:
$ docker ps
This command produces an output that looks like this:
CONTAINER ID IMAGE COMMAND ... PORTS NAMES 51a49fe8601a soumitag/multislide:2.0 "/bin/sh -c \"/usr/lo…" ... 0.0.0.0:8080->8080/tcp youthful_wozniak
The last column of the output shows the container name. In this example it is "youthful_wozniak"
To stop this container use
$ docker stop youthful_wozniak
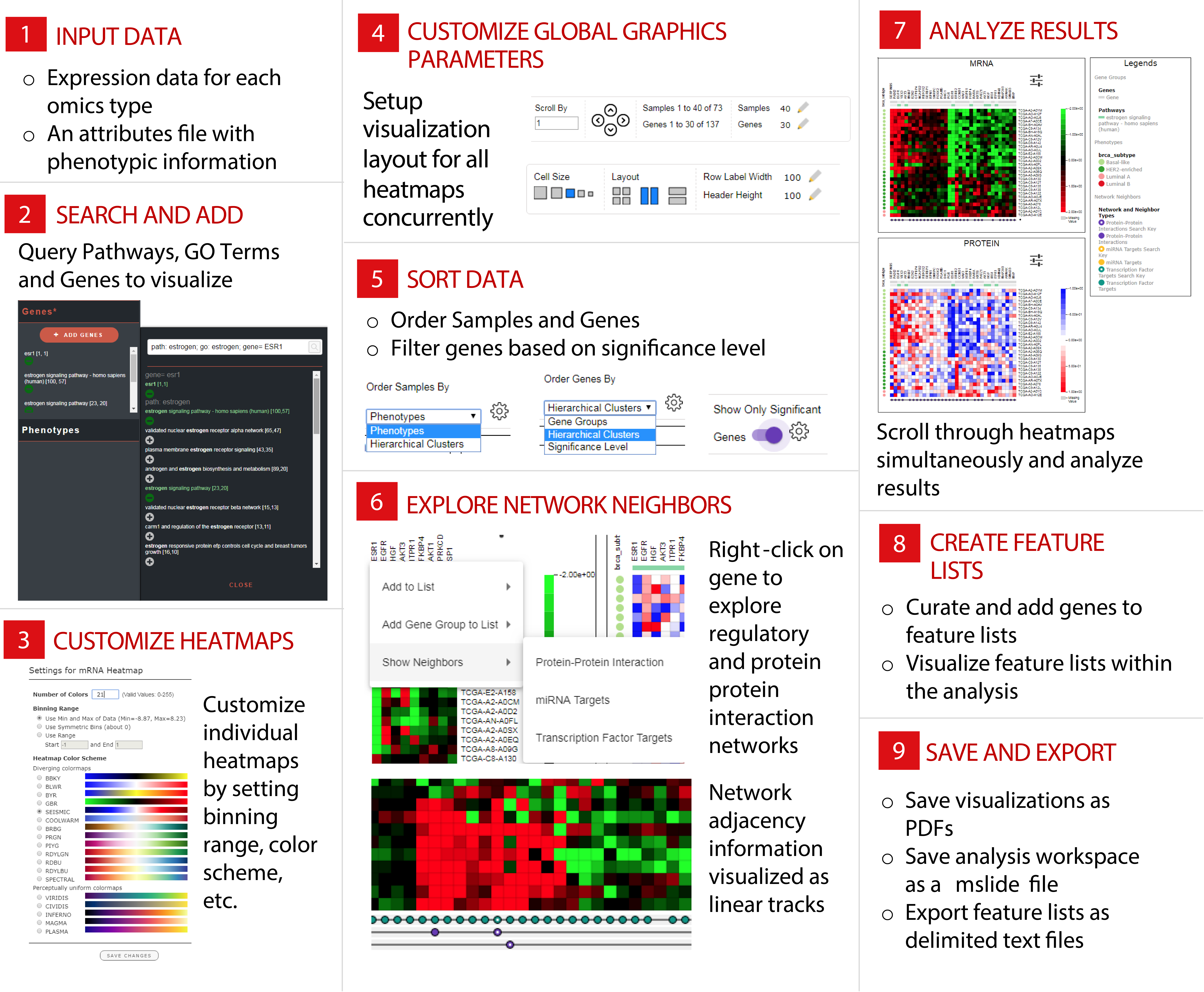
multiSLIDE is an open-source tool for query-driven visualization of quantitative single- or multi-omics data. Using pathways and networks as the basis for data linkage, multiSLIDE provides an interactive platform for querying the multi-omics data by genes, pathways, and intermolecular relationships.
- How to use multiSLIDE ?
- Videos tutorials demonstrating multiSLIDE's functionalities are available here