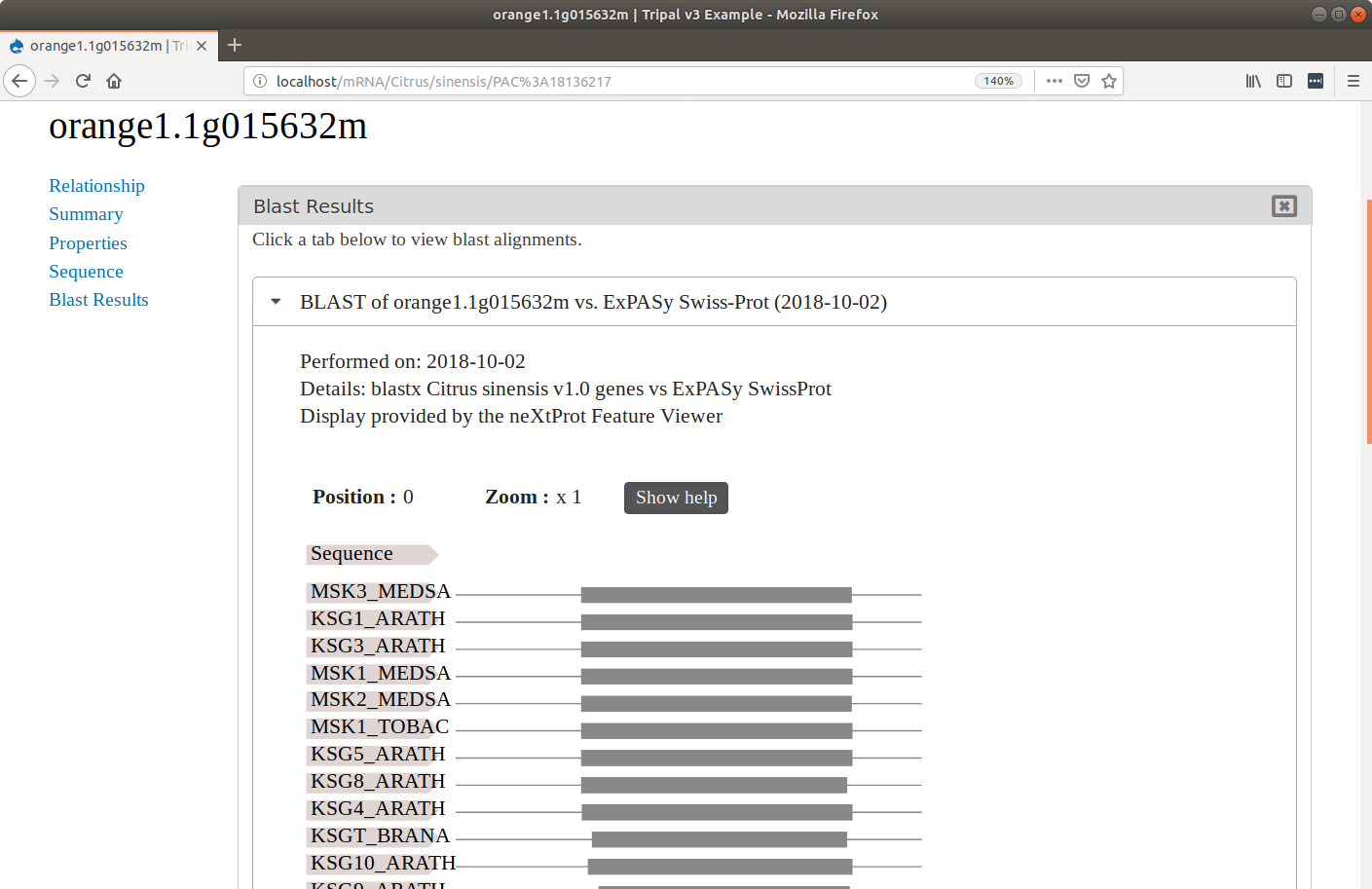
The Tripal Analysis BLAST module extends Tripal for display of BLAST local alignments on gene, mRNA or protein pages on a Tripal site. This module is compatible with both Tripal v2 and Tripal v3. It provides:
- An importer for loading BLAST XML results created using the NCBI blast program. Data is loaded into Chado and BLAST results are automatically associated with genes, mRNA (transcripts) or proteins.
- Blast results appear on each feature page via an interactive viewer and a tabular list (see screenshot for example).
In addition to display of BLAST results there are a few other modules that support BLAST. These include:
- Tripal Sequence Similarity Search. This module provides a web front-end for site visitors to execute BLAST within the Tripal site. It supports fast BLAST using Diamond and supports execution of BLAST jobs on a remote server (not the web server).
- Tripal BLAST. This module provides a web front-end for site visitors to execute BLAST within the Tripal site. It integrates with the Tripal Deamon (built into Tripal v3) module for fast execution of BLAST.
How is this module different? This module does not provide an interface for end-users to execute BLAST jobs. Rather it is meant to import BLAST results to appear on gene, transcript or protein pages to provide functional annotations.
Installation and usage instructionsare a provided in the Tripal User's Guide.