This is an R package for performing MultiSTAAR procedure in whole-genome sequencing studies.
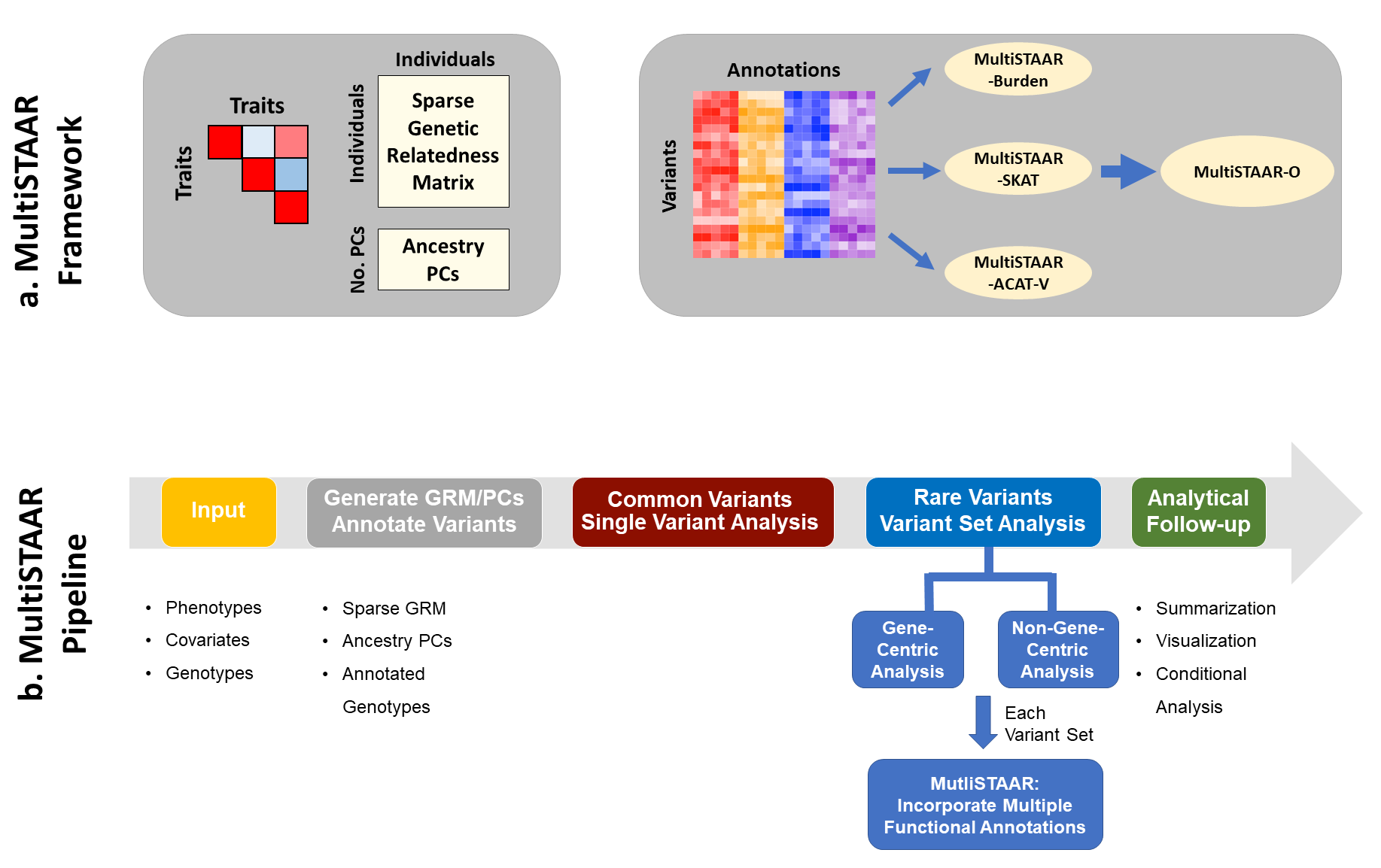
MultiSTAAR is an R package for performing Multi-trait variant-Set Test for Association using Annotation infoRmation (MultiSTAAR) procedure in whole-genome sequencing (WGS) studies. MultiSTAAR is a general framework that (1) leverages the correlation structure between multiple phenotypes to improve power of multi-trait analysis over single-trait analysis, and (2) incorporates both qualitative functional categories and quantitative complementary functional annotations using an omnibus multi-dimensional weighting scheme. MultiSTAAR accounts for population structure and relatedness, and is scalable for jointly analyzing large WGS studies of multiple correlated traits.
R (recommended version >= 3.5.1)
For optimal computational performance, it is recommended to use an R version configured with the Intel Math Kernel Library (or other fast BLAS/LAPACK libraries). See the instructions on building R with Intel MKL.
MultiSTAAR links to R packages Rcpp, RcppArmadillo and STAAR, and also imports R packages Rcpp, GMMAT, GENESIS, STAAR, Matrix. These dependencies should be installed before installing MultiSTAAR.
library(devtools)
devtools::install_github("xihaoli/MultiSTAAR",ref="main")
A docker image for MultiSTAAR, including R (version 3.6.1) built with Intel MKL and all STAAR-related packages (STAAR, MultiSTAAR, SCANG, STAARpipeline, STAARpipelineSummary) pre-installed, is located in the Docker Hub. The docker image can be pulled using
docker pull zilinli/staarpipeline:0.9.7
Please see the MultiSTAAR user manual for detailed usage of MultiSTAAR package. Please see the MultiSTAAR tutorial for an example of analyzing sequencing data using MultiSTAAR procedure. Please see the STAARpipeline tutorial for a detailed example of analyzing sequencing data using MultiSTAAR.
The whole-genome functional annotation data assembled from a variety of sources and the precomputed annotation principal components are available at the Functional Annotation of Variant - Online Resource (FAVOR) site and FAVOR Essential Database.
The current version is 0.9.7.1 (November 14, 2024).
This software is licensed under GPLv3.