Nikhil Keetha
·
Jay Karhade
·
Krishna Murthy Jatavallabhula
·
Gengshan Yang
·
Sebastian Scherer
Deva Ramanan
·
Jonathon Luiten
Paper | Video | Project Page
Table of Contents
SplaTAM has been tested on python 3.10, CUDA>=11.6. The simplest way to install all dependences is to use anaconda and pip in the following steps:
conda create -n splatam python=3.10
conda activate splatam
conda install -c "nvidia/label/cuda-11.6.0" cuda-toolkit
conda install pytorch==1.12.1 torchvision==0.13.1 torchaudio==0.12.1 cudatoolkit=11.6 -c pytorch -c conda-forge
pip install -r requirements.txtAlternatively, we also provide a conda environment.yml file :
conda env create -f environment.yml
conda activate splatamFor installation on Windows using Git bash, please refer to the instructions shared in Issue#9.
We also provide a docker image. We recommend using a venv to run the code inside a docker image:
docker pull nkeetha/splatam:v1
bash bash_scripts/docker_start.bash
cd /SplaTAM/
pip install virtualenv --user
mkdir venv
cd venv
virtualenv splatam --system-site-packages
pip install -r venv_requirements.txtSetting up a singularity container is similar:
cd </path/to/singularity/folder/
singularity pull splatam.sif docker://nkeetha/splatam:v1
singularity instance start --nv splatam.sif splatam
singularity run --nv instance://gradslam_2
cd <path/to/SplaTAM/>
pip install virtualenv --user
mkdir venv
cd venv
virtualenv splatam --system-site-packages
pip install -r venv_requirements.txtYou can SplaTAM your own environment with an iPhone or LiDAR-equipped Apple device by downloading and using the NeRFCapture app.
Make sure that your iPhone and PC are connected to the same WiFi network, and then run the following command:
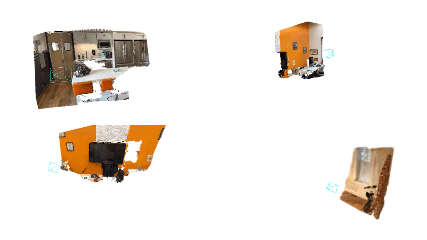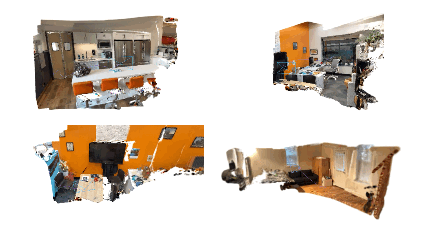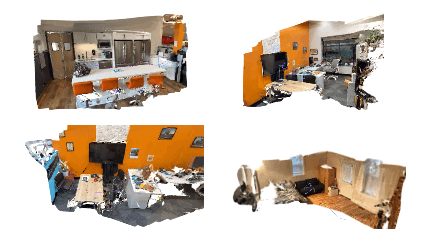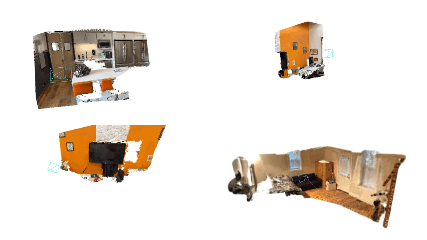
bash bash_scripts/online_demo.bash configs/iphone/online_demo.pyOn the app, keep clicking send for successive frames. Once the capturing of frames is done, the app will disconnect from the PC and check out SplaTAM's interactive rendering of the reconstruction on your PC! Here are some cool example results:
You can also first capture the dataset and then run SplaTAM offline on the dataset with the following command:
bash bash_scripts/nerfcapture.bash configs/iphone/nerfcapture.pyIf you would like to only capture your own iPhone dataset using the NeRFCapture app, please use the following command:
bash bash_scripts/nerfcapture2dataset.bash configs/iphone/dataset.pyWe will use the iPhone dataset as an example to show how to use SplaTAM. The following steps are similar for other datasets.
To run SplaTAM, please use the following command:
python scripts/splatam.py configs/iphone/splatam.pyTo visualize the final interactive SplaTAM reconstruction, please use the following command:
python viz_scripts/final_recon.py configs/iphone/splatam.pyTo visualize the SplaTAM reconstruction in an online fashion, please use the following command:
python viz_scripts/online_recon.py configs/iphone/splatam.pyTo run 3D Gaussian Splatting on the SplaTAM reconstruction, please use the following command:
python scripts/post_splatam_opt.pt configs/iphone/post_splatam_opt.pyTo run 3D Gaussian Splatting on a dataset using ground truth poses, please use the following command:
python scripts/gaussian_splatting.py configs/iphone/gaussian_splatting.pyDATAROOT is ./data by default. Please change the input_folder path in the scene-specific config files if datasets are stored somewhere else on your machine.
Download the data as below, and the data is saved into the ./data/Replica folder. Note that the Replica data is generated by the authors of iMAP (but hosted by the authors of NICE-SLAM). Please cite iMAP if you use the data.
bash bash_scripts/download_replica.shbash bash_scripts/download_tum.shPlease follow the data downloading procedure on the ScanNet website, and extract color/depth frames from the .sens file using this code.
[Directory structure of ScanNet (click to expand)]
DATAROOT
└── scannet
└── scene0000_00
└── frames
├── color
│ ├── 0.jpg
│ ├── 1.jpg
│ ├── ...
│ └── ...
├── depth
│ ├── 0.png
│ ├── 1.png
│ ├── ...
│ └── ...
├── intrinsic
└── pose
├── 0.txt
├── 1.txt
├── ...
└── ...
We use the following sequences:
scene0000_00
scene0059_00
scene0106_00
scene0181_00
scene0207_00
Please follow the data downloading and image undistortion procedure on the ScanNet++ website. Additionally for undistorting the DSLR depth images, we use our own variant of the official ScanNet++ processing code. We will open a pull request to the official ScanNet++ repository soon.
We use the following sequences:
8b5caf3398
b20a261fdf
For b20a261fdf, we use the first 360 frames, due to an abrupt jump/teleportation in the trajectory post frame 360. Please note that ScanNet++ was primarily intended as a NeRF Training & Novel View Synthesis dataset.
We use the Replica-V2 dataset from vMAP to evaluate novel view synthesis. Please download the pre-generated replica sequences from vMAP.
For running SplaTAM, we recommend using weights and biases for the logging. This can be turned on by setting the wandb flag to True in the configs file. Also make sure to specify the path wandb_folder. If you don't have a wandb account, first create one. Please make sure to change the entity config to your wandb account. Each scene has a config folder, where the input_folder and output paths need to be specified.
Below, we show some example run commands for one scene from each dataset. After SLAM, the trajectory error will be evaluated along with the rendering metrics. The results will be saved to ./experiments by default.
To run SplaTAM on the room0 scene, run the following command:
python scripts/splatam.py configs/replica/splatam.pyTo run SplaTAM-S on the room0 scene, run the following command:
python scripts/splatam.py configs/replica/splatam_s.pyFor other scenes, please modify the configs/replica/splatam.py file or use configs/replica/replica.bash.
To run SplaTAM on the freiburg1_desk scene, run the following command:
python scripts/splatam.py configs/tum/splatam.pyFor other scenes, please modify the configs/tum/splatam.py file or use configs/tum/tum.bash.
To run SplaTAM on the scene0000_00 scene, run the following command:
python scripts/splatam.py configs/scannet/splatam.pyFor other scenes, please modify the configs/scannet/splatam.py file or use configs/scannet/scannet.bash.
To run SplaTAM on the 8b5caf3398 scene, run the following command:
python scripts/splatam.py configs/scannetpp/splatam.pyTo run Novel View Synthesis on the 8b5caf3398 scene, run the following command:
python scripts/eval_novel_view.py configs/scannetpp/eval_novel_view.pyFor other scenes, please modify the configs/scannetpp/splatam.py file or use configs/scannetpp/scannetpp.bash.
To run SplaTAM on the room0 scene, run the following command:
python scripts/splatam.py configs/replica_v2/splatam.pyTo run Novel View Synthesis on the room0 scene post SplaTAM, run the following command:
python scripts/eval_novel_view.py configs/replica_v2/eval_novel_view.pyFor other scenes, please modify the config files.
We thank the authors of the following repositories for their open-source code:
- 3D Gaussians
- Dataloaders
- Baselines
If you find our paper and code useful, please cite us:
@article{keetha2023splatam,
author = {Keetha, Nikhil and Karhade, Jay and Jatavallabhula, Krishna Murthy and Yang, Gengshan and Scherer, Sebastian and Ramanan, Deva and Luiten, Jonathan}
title = {SplaTAM: Splat, Track & Map 3D Gaussians for Dense RGB-D SLAM},
journal = {arXiv},
year = {2023},
}