Protein structure handling in Python made easy.
Dealing with a batch of protein structures is a common task in this new era of artificial intelligence-based protein structure prediction, optimization, and design.
This package, protstruc, provides an easy-to-use interface to handle a batch of protein structures, while specifically designed for deep learning applications based on PyTorch.
protstruc provides various features including:
-
Computing geometric features of a protein structure
- pairwise distance matrix
- backbone dihedrals
- backbone orientations
- backbone translations
- pairwise dihedrals
- pairwise planar angles
- Manipulation of protein structures
- selecting specific chains
- selecting specific residues
- rotating the structure
- translating the structure
- zeroing center-of-mass
- diffusing (adding Gaussian noise) atom 3D coordinates
There are many options to initialize the core data structure for a batch of protein structures, StructureBatch:
- from local PDB files
StructureBatch.from_pdb - from PDB IDs
StructureBatch.from_pdb_id - Backbone (or even full-) atom 3D coordinates
StructureBatch.from_xyz - Backbone orientation and translations
StructureBatch.from_backbone_orientations_translations - Dihedral angles
StructureBatch.from_dihedrals(WIP)
Simple enough, just use pip:
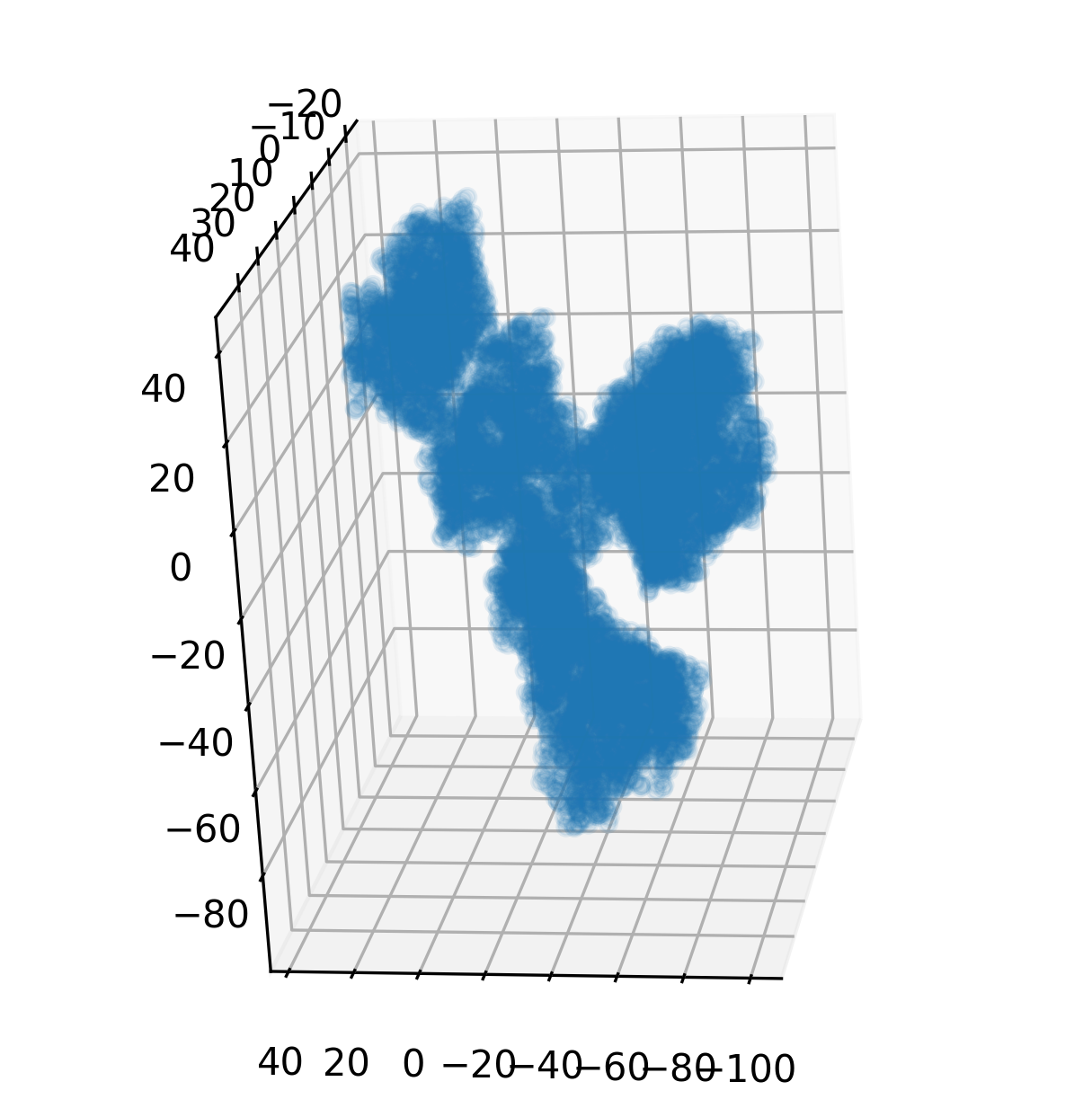
pip install protstrucThis code snippet loads a structure of a renowned monoclonal antibody targeting PD-1, named Pembrolizumab, from its PDB identifier 5DK3, and visualizes its structure.
import matplotlib.pyplot as plt
from protstruc import StructureBatch
# Load a structure from a PDB identifier
struc = StructureBatch.from_pdb_id('5dk3')
# Full-atom 3D coordinates of shape (batch_size, num_residues, max_num_atoms_per_residue, 3)
# Note that max_num_atoms_per_residue is set to 15 by default.
# Also note that in this case, batch_size=1
xyz = struc.get_xyz()
# Visualize the structure
fig = plt.figure(figsize=(5, 5))
ax = fig.add_subplot(111, projection='3d')
ax.scatter(
xyz[0, :, :, 0], xyz[0, :, :, 1], xyz[0, :, :, 2],
alpha=0.1, c='C0'
)
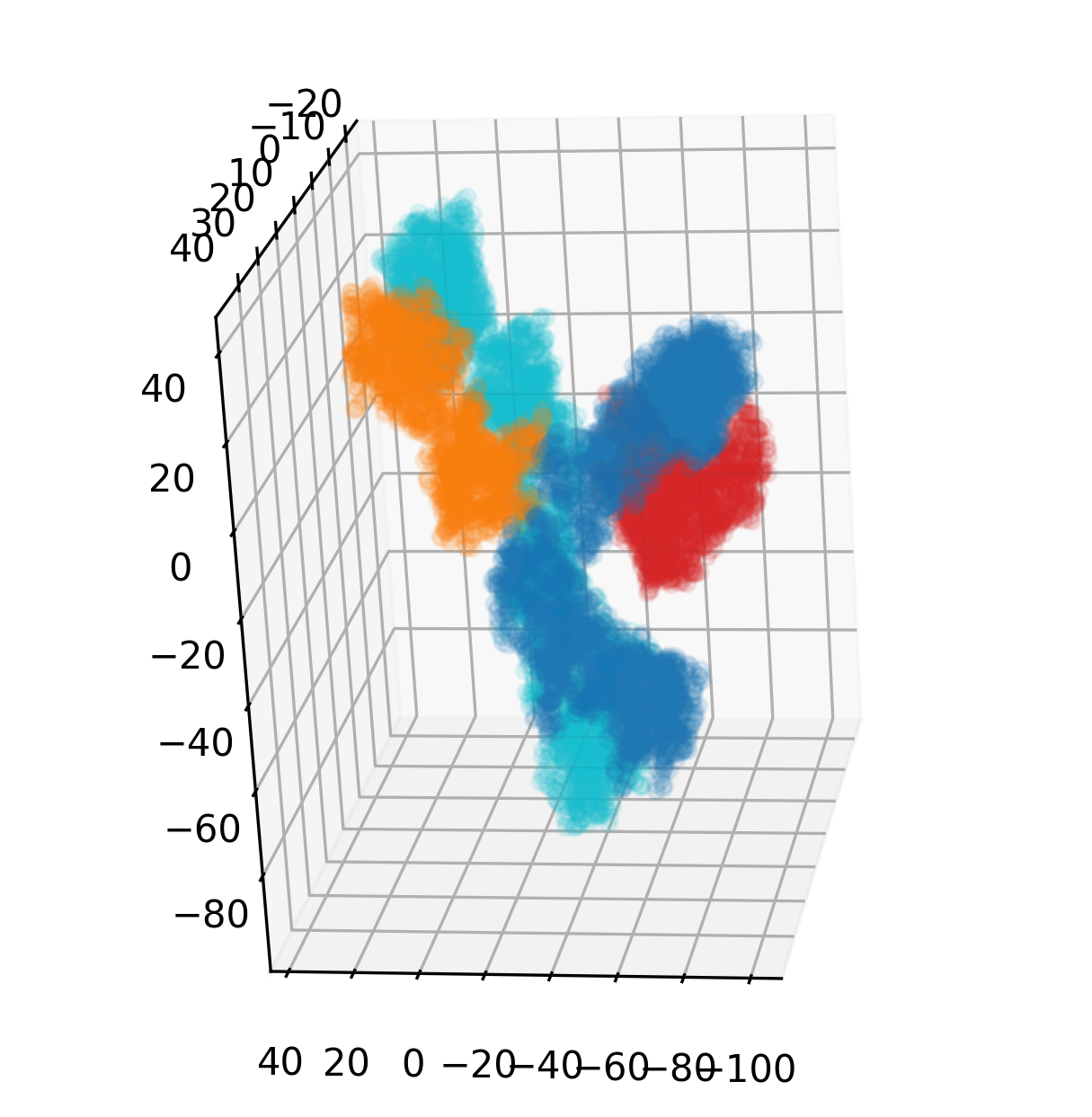
ax.view_init(20, 20)You may need a selector for specific peptide chains for some reason.
A StructureBatch object provides handy methods get_chain_ids and get_chain_idx to get chain IDs and the integer indices to represent the corresponding chains, respectively.
The following code snippet generates boolean masks for each chain, uses them to select specific chain and colors heavy chain with bluish colors and light chain with reddish colors.
chain_idx = struc.get_chain_idx()[0] # Note we only have one structure in the batch.
chain_ids = struc.get_chain_ids()[0]
# Let heavy chains colored in bluish colors,
# and light chains colored in reddish ones.
colors = ['C3', 'C0', 'C1', 'C9']
for idx, (chain_id, c) in enumerate(zip(chain_ids, colors)):
mask = chain_idx == idx
ax.scatter(
xyz[0, mask, :, 0], xyz[0, mask, :, 1], xyz[0, mask, :, 2],
alpha=0.1, c=c
)
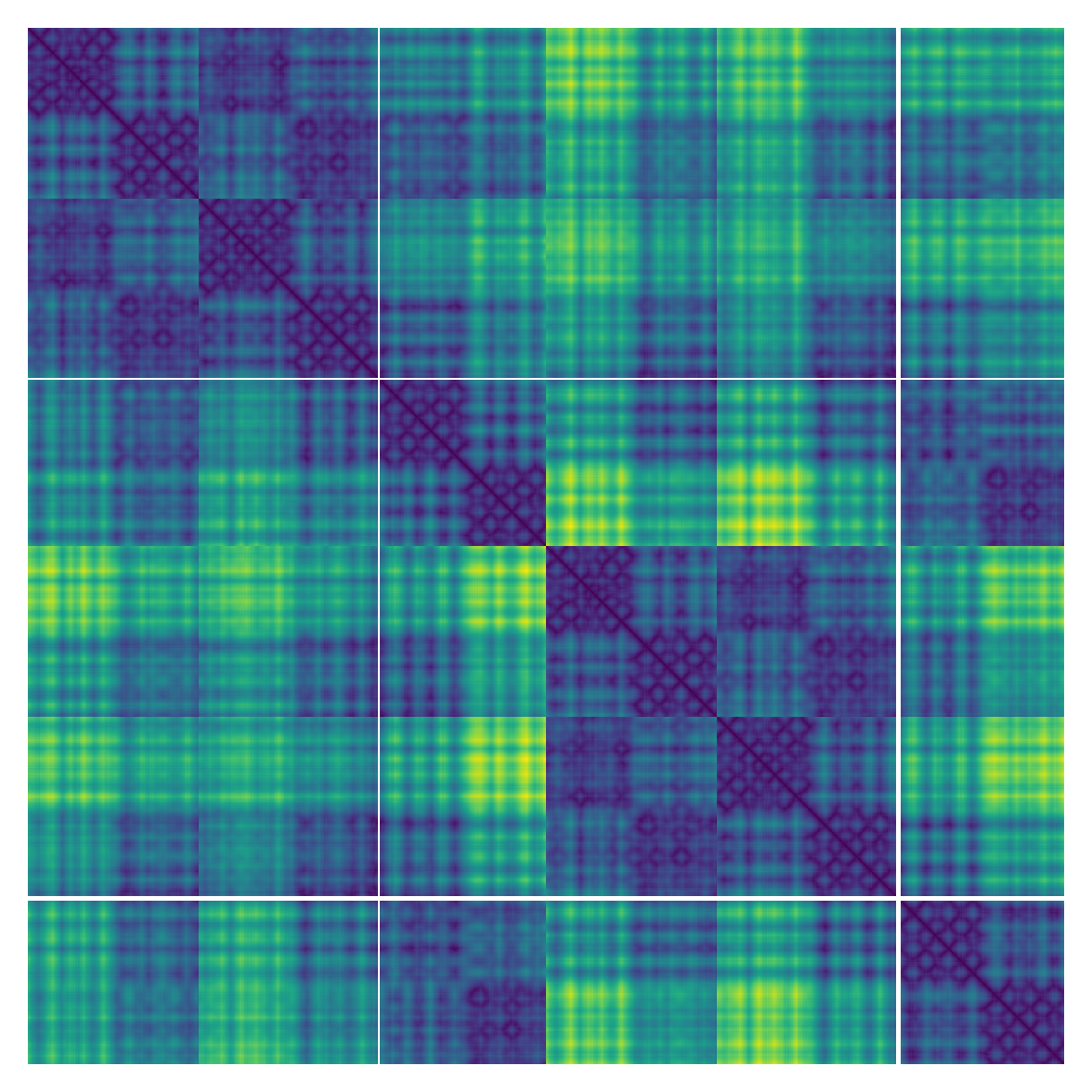
ax.view_init(-10, 20, 90)Pairwise distance matrix
from protstruc.general import ATOM
struc = StructureBatch.from_pdb_id('5dk3')
# Shape (batch_size, num_residue, num_residue, num_atom, num_atom)
distmat, distmat_mask = struc.pairwise_distance_matrix()
plt.matshow(distmat[0, :, :, ATOM.CA, ATOM.CA]) # Ca-Ca distance matrix
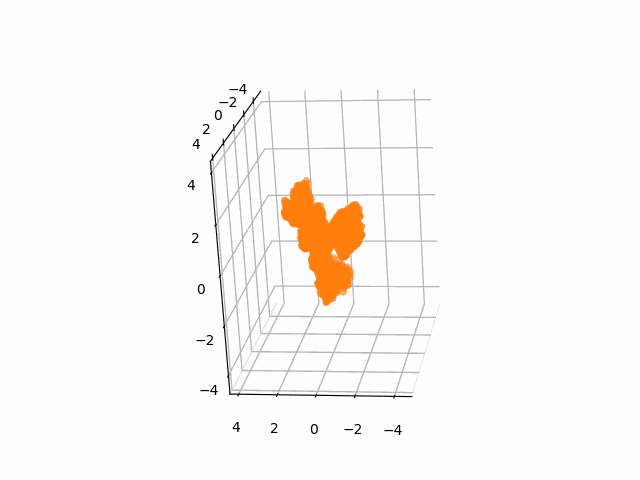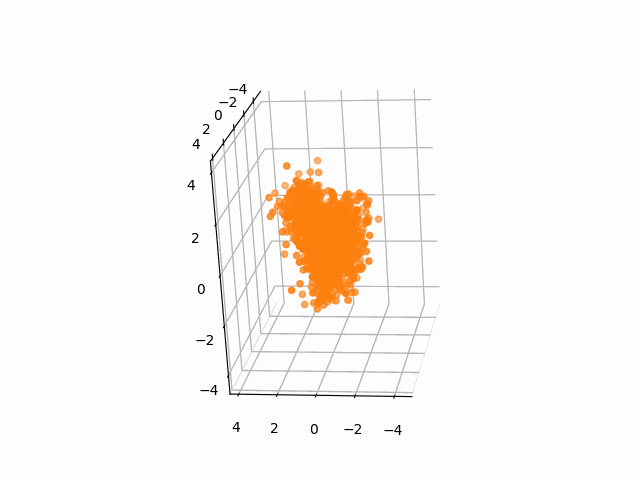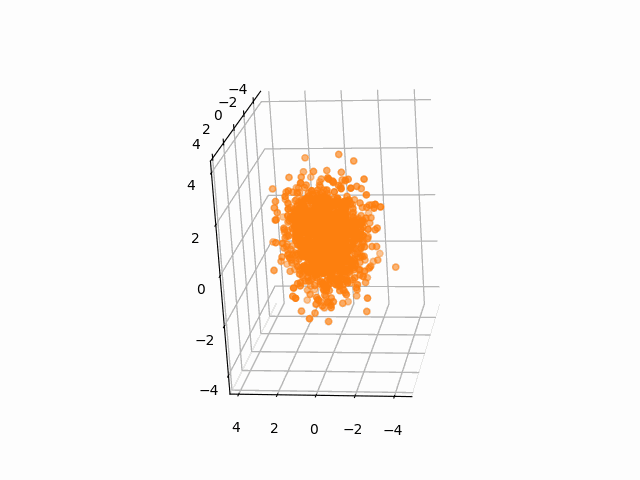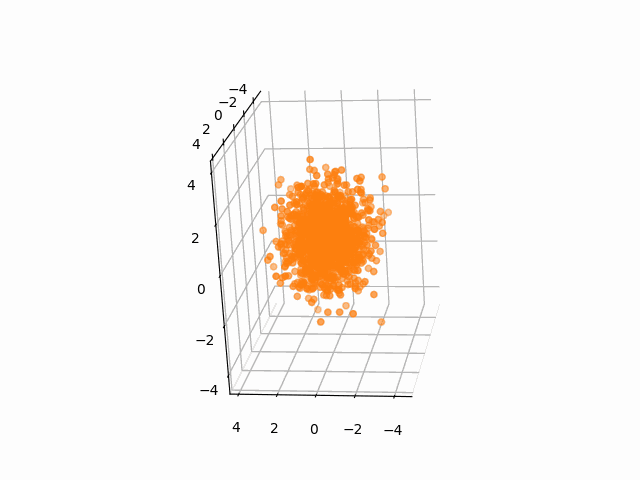
plt.axis('off')We often need to add Gaussian noises to 3D atom coordinates to train Euclidean diffusion models for protein structure generation.
A StructureBatch object provides a handy method diffuse_xyz to add Gaussian noises of zero mean and given variances to 3D atom coordinates.
# Diffusion parameters
T = 300
sched = cosine_variance_schedule(T)
prt_idx = 0
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
struc = StructureBatch.from_pdb_id('5dk3')
# Center atom coordinates at the origin and scale to unit variance
# NOTE: All atoms are considered here
struc.standardize()
ims = []
for t in range(T):
xyz = struc.get_xyz()
im1 = ax.scatter(
xyz[prt_idx, :, ATOM.CA, 0],
xyz[prt_idx, :, ATOM.CA, 1],
xyz[prt_idx, :, ATOM.CA, 2],
c='C1'
)
ax.view_init(-10, 20, 90)
ims.append([im1])
# Add Gaussian noise to the structure
struc.diffuse_xyz(beta=torch.tensor([sched['beta'][t]]))
ani = animation.ArtistAnimation(fig, ims, interval=100, blit=True, repeat_delay=1000)
ani.save(f'animations/pembrolizumab_diffusion.gif')To test the integrity of the package, run the following command:
pytest