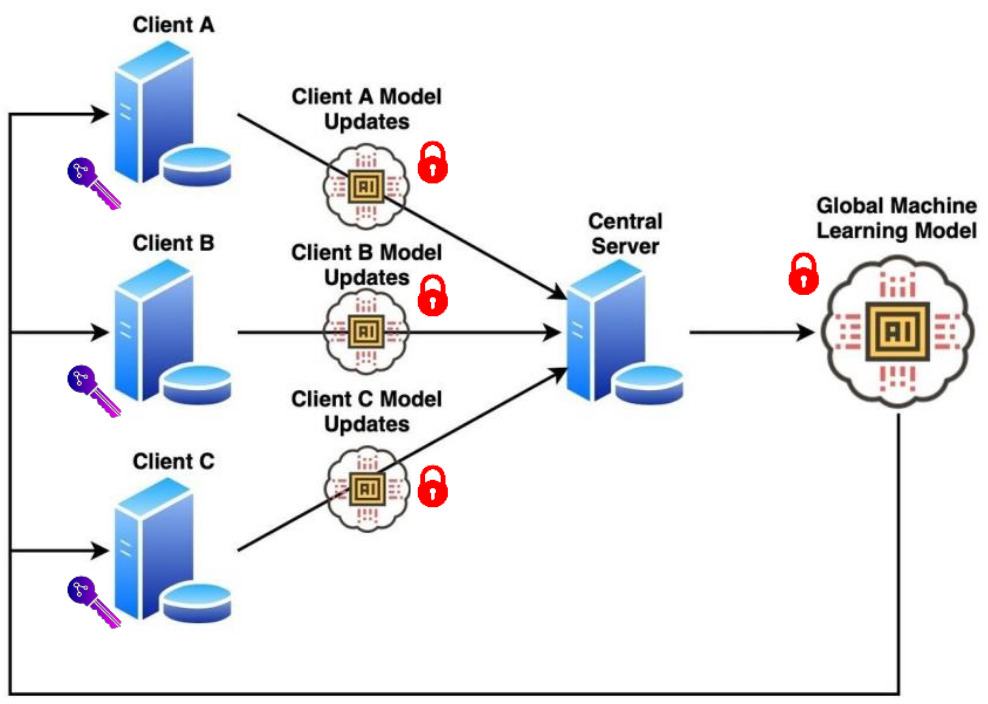
- This is the official repository of the paper Encrypted federated learning for secure decentralized collaboration in cancer image analysis.
Federated learning with homomorphic encryption enables multiple parties to securely co-train artificial intelligence models in pathology and radiology, reaching state-of-the-art performance with privacy guarantees.
The software is developed in Python 3.8. For the deep learning, the PyTorch 1.4 framework is used. The secure federated learning process was developed using PySyft 0.2.9.
Main Python modules required for the software can be installed from ./requirements in three stages:
- Create a Python3 environment by installing the conda
environment.ymlfile:
$ conda env create -f environment.yml
$ source activate SHEFL
$ pip install syft==0.2.9
- Install the remaining dependencies from
requirements.txt.
Note: These might take a few minutes.
Our source code for secure federated learning using homomorphic encryption for both the radiology and pathology parts as well as training and evaluation of the deep neural networks, MR image analysis and preprocessing, 3D data augmentation, and gradient inversion attack are available here.
- Everything for the radiology can be run from ./main_3D_brats.py.
- The data preprocessing parameters, directories, hyper-parameters, and model parameters can be modified from ./configs/config.yaml.
- Also, you should first choose an
experimentname (if you are starting a new experiment) for training, in which all the evaluation and loss value statistics, tensorboard events, and model & checkpoints will be stored. Furthermore, aconfig.yamlfile will be created for each experiment storing all the information needed. - For testing, just load the experiment which its model you need.
- The rest of the files:
- ./models/ directory contains all the model architectures and losses.
- ./data/ directory contains all the data preprocessing, 3D augmentation, and loading files.
- ./Train_Valid_brats.py contains the training and validation processes of radiology.
- ./gradient_inversion/ directory contains all the files for gradient inversion attack.
- ./pathology/ directory contains all the files for the pathology part. The exact same structure exists in ./pathology/ as for the radiology part. Everything for the pathology can be run from ./main_pathology.py. ./Train_Valid_pathology.py contains the training and validation processes of pathology. ./Prediction_pathology.py* contains the evaluation process of pathology.
Note: All source codes for the histological image analysis and histological image preprocessing are available at https://github.com/KatherLab.
The data that support the findings of this study are in part publicly available, in part proprietary datasets provided under collaboration agreements.
- BraTS20 dataset is public under: link.
- Data (including histological images) from the TCGA database are available at: link.
- All molecular data for patients in the TCGA cohorts are available at: link.
- Data access for the Northern Ireland Biobank can be requested at: link.
- All other data can be requested from the respective study groups who independently manage data access for their study cohorts.
Truhn D, Tayebi Arasteh S, et al. "Encrypted federated learning for secure decentralized collaboration in cancer image analysis". Medical Image Analysis, https://doi.org/10.1016/j.media.2023.103059, 2024.
@article {SHEFL2024,
author = {Truhn, Daniel and Tayebi Arasteh, Soroosh and Saldanha, Oliver Lester and Mueller-Franzes, Gustav and Khader, Firas and Quirke, Philip and West, Nicholas P. and Gray, Richard and Hutchins, Gordon G. A. and James, Jacqueline A. and Loughrey, Maurice B. and Salto-Tellez, Manuel and Brenner, Hermann and Brobeil, Alexander and Yuan, Tanwei and Chang-Claude, Jenny and Hoffmeister, Michael and Foersch, Sebastian and Han, Tianyu and Keil, Sebastian and Schulze-Hagen, Maximilian and Isfort, Peter and Bruners, Philipp and Kaissis, Georgios and Kuhl, Christiane and Nebelung, Sven and Kather, Jakob Nikolas},
title = {Encrypted federated learning for secure decentralized collaboration in cancer image analysis},
year = {2024},
doi = {10.1016/j.media.2023.103059},
URL = {https://doi.org/10.1016/j.media.2023.103059},
journal = {Medical Image Analysis}
}