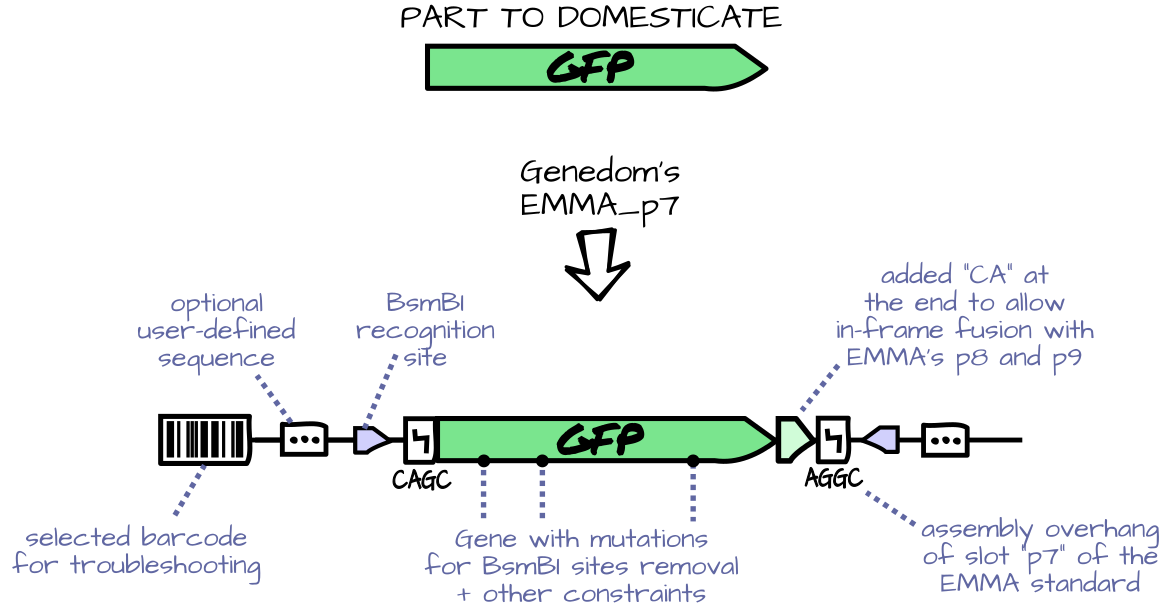
GeneDom is a python library for managing the domestication of genetic parts (i.e. the modification of their sequence so as to make them compatible with a given genetic assembly standard). Genedom binds together a sequence optimizer, genetic standards informations, and a reporting routine, to automate the domestication of large batches in an easy and human-friendly way.
Features include:
- Possibility to define part domesticators with added right-hand and left-hand nucleotide, hard constraints on the sequence (such as absence of a restriction site) and optimization objectives (such as codon optimization).
- Built-in pre-defined domesticators for popular genetic assembly standards (well, only EMMA at the moment).
- Possibility to generate and attribute barcodes that will be added to the sequence (but won't be in final constructs) in order to easily check that this is the right part in the future in case of label mix-up.
- Routine for mass-domesticating sequences with report generation, including reports on each sequence optimization, spreadsheets of parts, ready-to-order FASTA records of the parts, and a summary report to quickly verify everything, with a list of every domesticator used, for traceability.
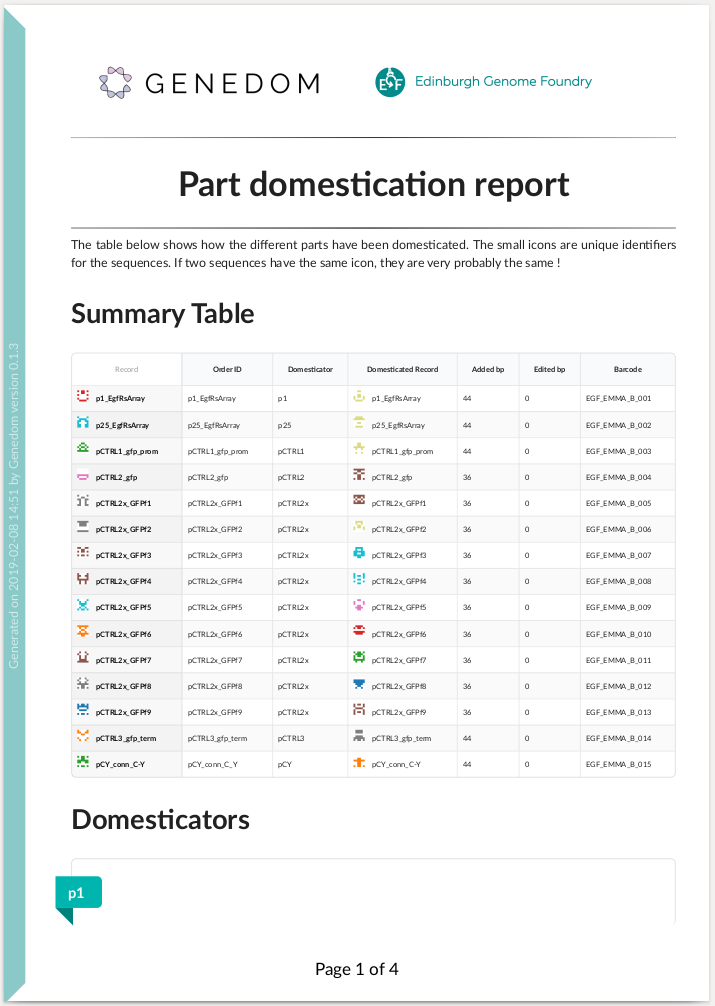
Here is an example of summary report:
You can also use Genedom online via this web app.
from genedom import (GoldenGateDomesticator, random_dna_sequence,
write_record)
sequence = random_dna_sequence(2000, seed=123)
domesticator = GoldenGateDomesticator("ATTC", "ATCG", enzyme='BsmBI')
domestication_results = domesticator.domesticate(sequence, edit=True)
print (domestication_results.summary())
write_record(domestication_results.record_after, 'domesticated.gb')(see docs for more potions)
from genedom import BarcodesCollection
barcodes_collection = BarcodesCollection.from_specs(
n_barcodes=96, barcode_length=20,
forbidden_enzymes=('BsaI', 'BsmBI', 'BbsI'))
barcodes_collection.to_fasta('example_barcodes_collection.fa')from genedom import BUILTIN_STANDARDS, load_record, batch_domestication
records = [
load_record(filepath, name=filename)
for filepath in records_filepaths
]
barcodes_collection = BarcodesCollection.from_specs(n_barcodes=10)
batch_domestication(records, 'domestication_report.zip',
barcodes=barcodes, # optional
standard=BUILTIN_STANDARDS.EMMA)You can install Genedom through PIP:
pip install genedomAlternatively, you can unzip the sources in a folder and type:
python setup.py installGenedom is an open-source software originally written at the Edinburgh Genome Foundry by Zulko and released on Github under the MIT license (Copyright 2018 Edinburgh Genome Foundry). Everyone is welcome to contribute !