🚀 The purpose of this document is to help you become productive as quickly as possible with the multiSight package.
- The goal of multiSight is to handle multi-omics data and network inference in a easy-to-use R shiny package.
You could use this tool with a graphical interface or only with script functions (see Vignette and manual for detailed examples).
You can install the released version of multiSight from Bioconductor with:
#To install this package ensure you have BiocManager installed
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
#The following initializes usage of Bioc devel
BiocManager::install("multiSight")multiSight is an R package providing an user-friendly graphical interface to analyze and explore your omic datasets in a multi-omics manner by DESeq2 (see Biological Insights tab), machine learning methods with biosigner and multi-block statistical analysis (see Classification tab) helped by p-values pooling Stouffer’s method.
Classification models are fitted to select few subsets of features, using biosigner or sPLS-DA methods. biosigner provides one model by omic block and one list of features named biosignature. Nevertheless, sPLS-DA biosignatures are based on more features than biosigner.
Biosignatures can be used:
- To forecast phenotype (e.g. for diagnostic, histological subtyping);
- To design Pathway and gene ontology enrichment (sPLS-DA biosignatures only);
- To build Network inference;
- To find PubMed references to make assumptions easier and data-driven.
multiSight enables you to get better biological insights for each omic dataset helping by four analytic modules which content:
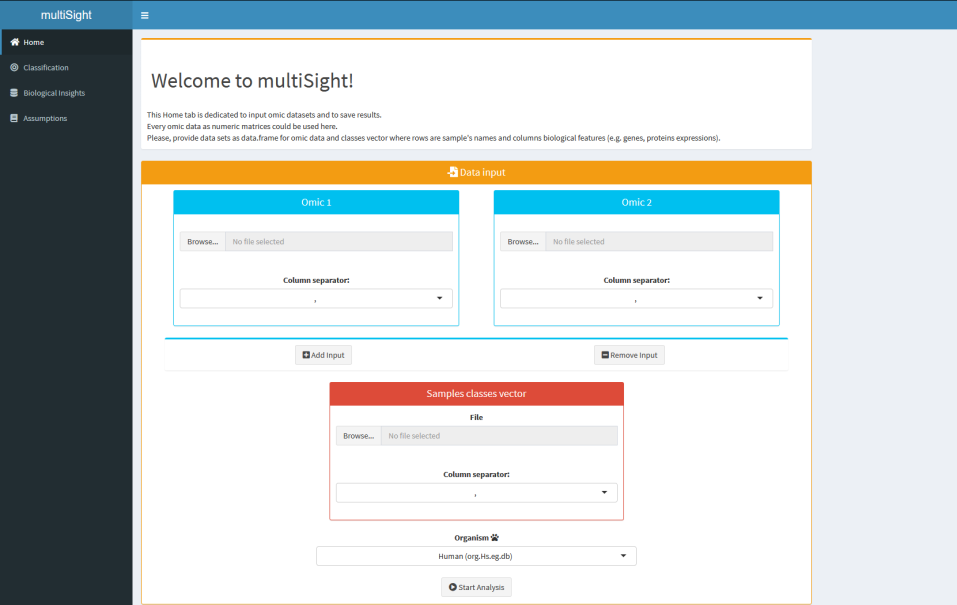
- 📝 Data input & results;
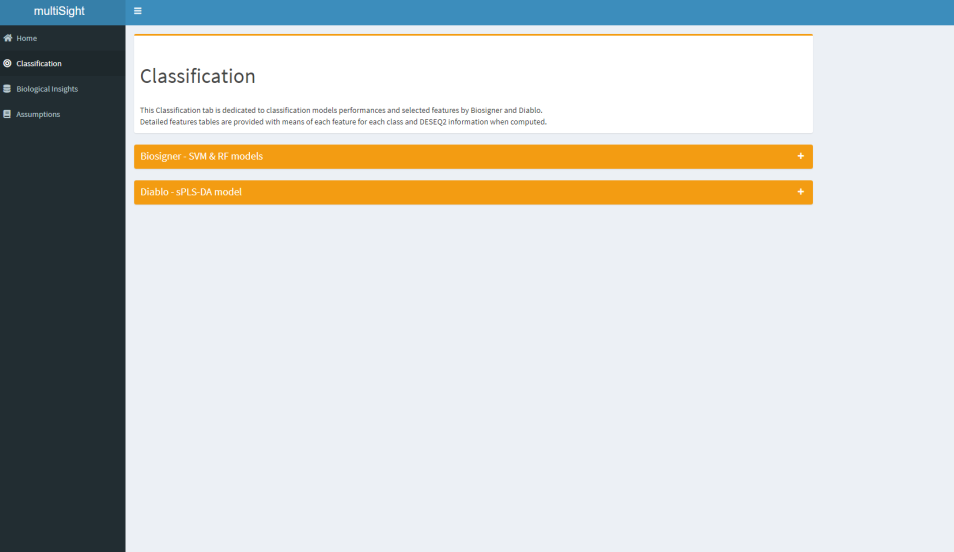
- 🎯 Classification models building;
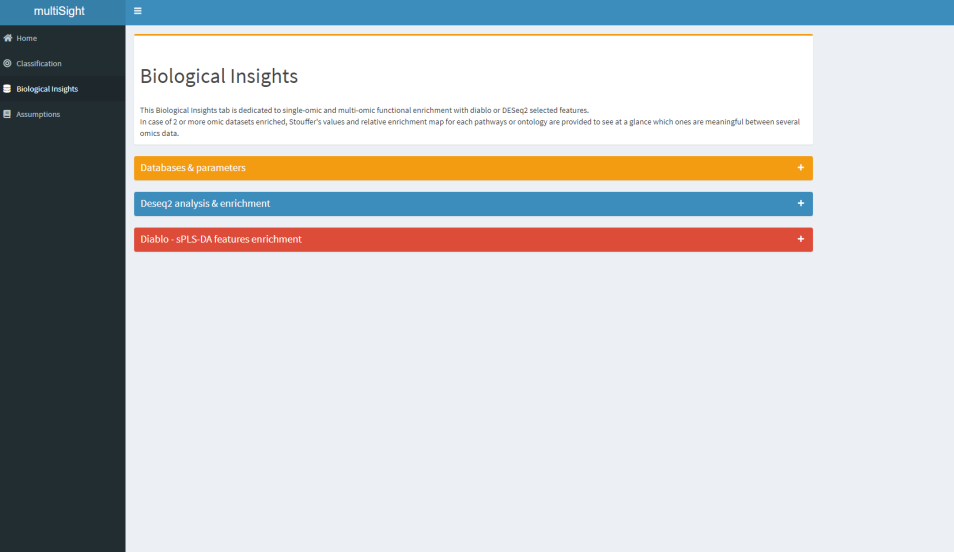
- 📚 Biological databases querying;
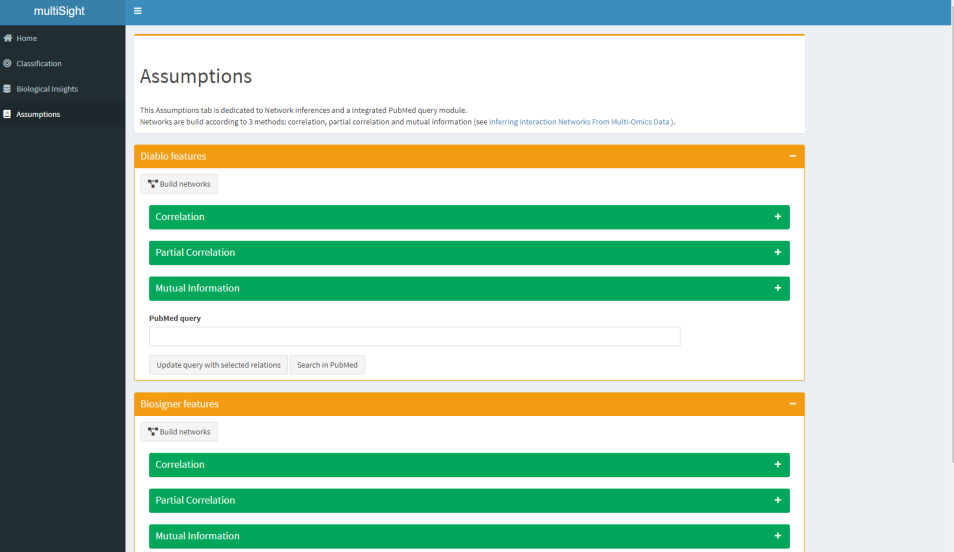
- 🌱 Network Inference & PubMed querying.
👉 Run the application
run_app()| 📝 Home | 🎯 Classification | 📚 Biological Insights | 🌱 Assumption |
|---|---|---|---|
 |
 |
 |
 |
All types of omic data respecting input format is supported to build classification models, biosignatures selection and network inference.
- Genomics;
- Transcriptomics;
- Proteomics;
- Metabolomics;
- Lipidomics;
👉 In fact all numeric matrices.
You have to provide two types of data: numeric matrices and classes vector as csv tables for all same samples.
| SIGIRR | MAOA | MANSC1 | ||
|---|---|---|---|---|
| AOFJ | 0 | 150 | 1004 | … |
| A13E | 34 | 0 | 0 | |
| … |
| ENSG00000139618 | ENSG00000226023 | ENSG00000198695 | ||
|---|---|---|---|---|
| AOFJ | 25 | 42 | 423 | … |
| A13E | 0 | 154 | 4900 | |
| … |
… 👉 unlimited number of omic datasets.
| 4292 | 5254 | 7432 | ||
|---|---|---|---|---|
| AOFJ | 25 | 42 | 423 | … |
| A13E | 0 | 154 | 4900 | |
| … |
| Y | |
|---|---|
| AOFJ | condA |
| A13E | condB |
| … |
Two types of models have been implemented so far to answer different questions: biosigner & sPLS-DA (DIABLO) .
- To determine small biosignatures - biosigner.
- To build classification models in a multi-omics way - DIABLO.
- To select relevant biological features to enrich - DIABLO.
| Features selected | Performances |
|---|---|
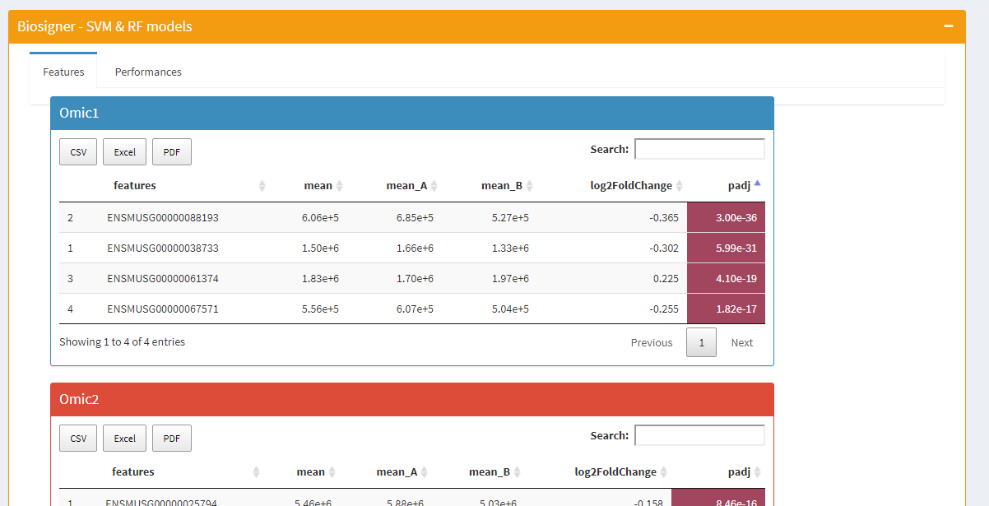 |
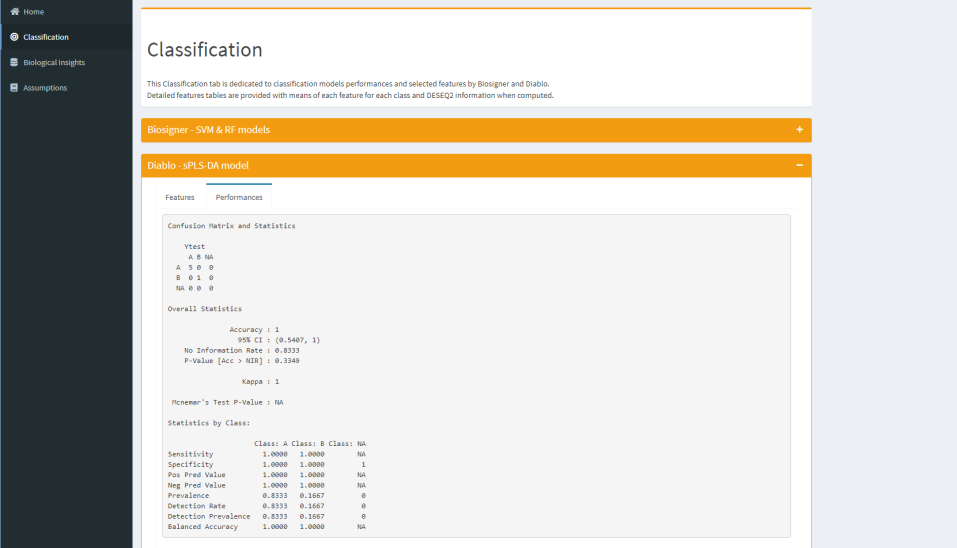 |
Biological Insight tab is dedicated to give biological sense to your data.
- You could process 2 analysis in 2 clicks: both DESeq2 and DIABLO features ORAs for functional enrichment.
multiSight uses so far several databases to provide a large panel of enrichment analysis, automatically after few clicks:
Pathways and Gene Ontology databases are implemented, helped by clusterProfiler and reactomePA R Bioconductor packages.
- Kegg;
- Reactome;
- wikiPathways;
- Molecular Function (GO)
- Cellular Component (GO)
- Biological Process (GO)
Two types of result visualization are given:
- Classical Enrichment tables for each omic and each database (e.g. Pathways id, p-value, padjust columns).
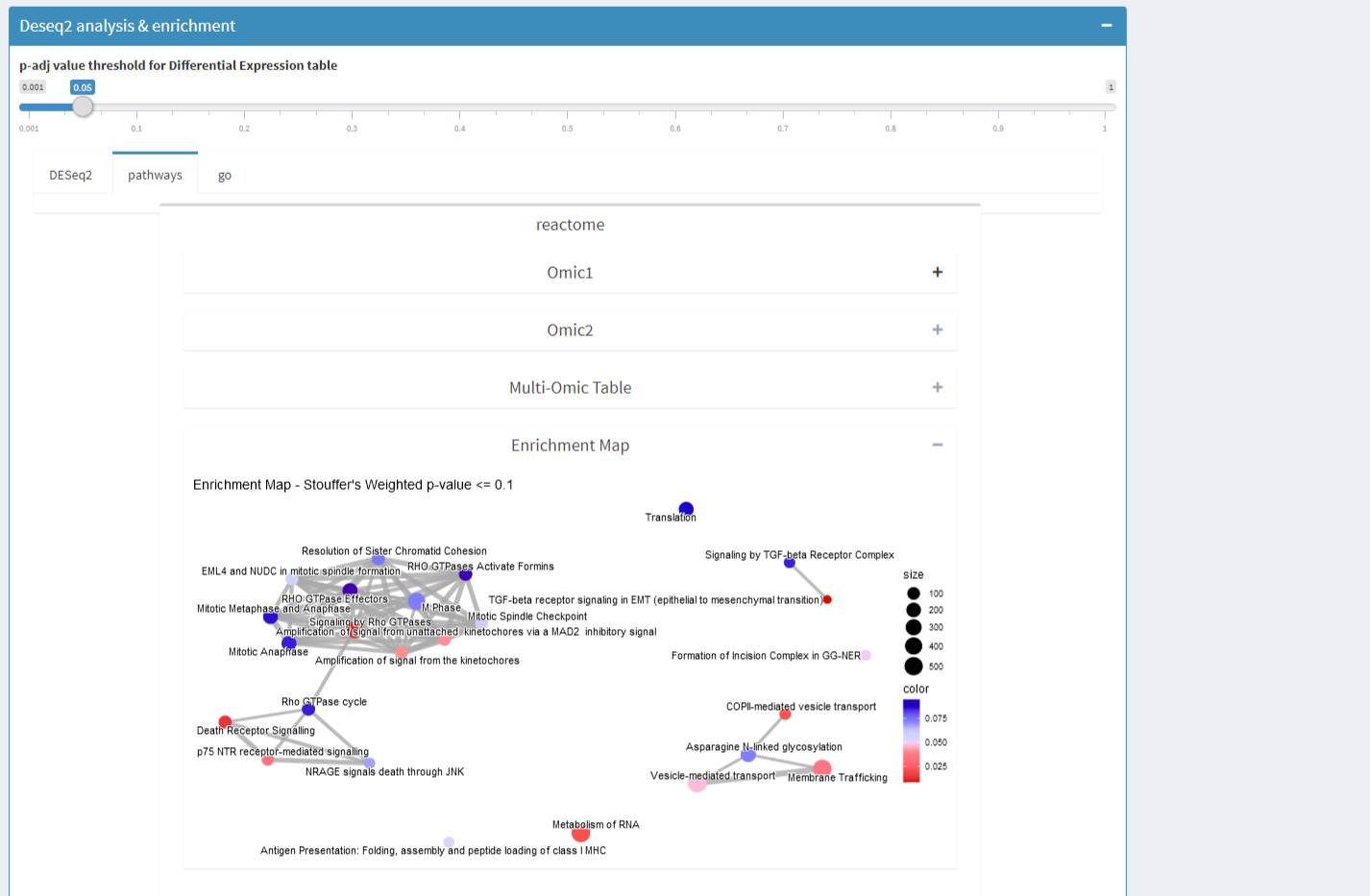
- And, when more than one omic enriched: a Multi-omics table and a multi-omics enrichment map for DESeq2 and DIABLO selected features.
| DESeq2 & DIABLO features | Enrichment tables | Enrichment Map |
|---|---|---|
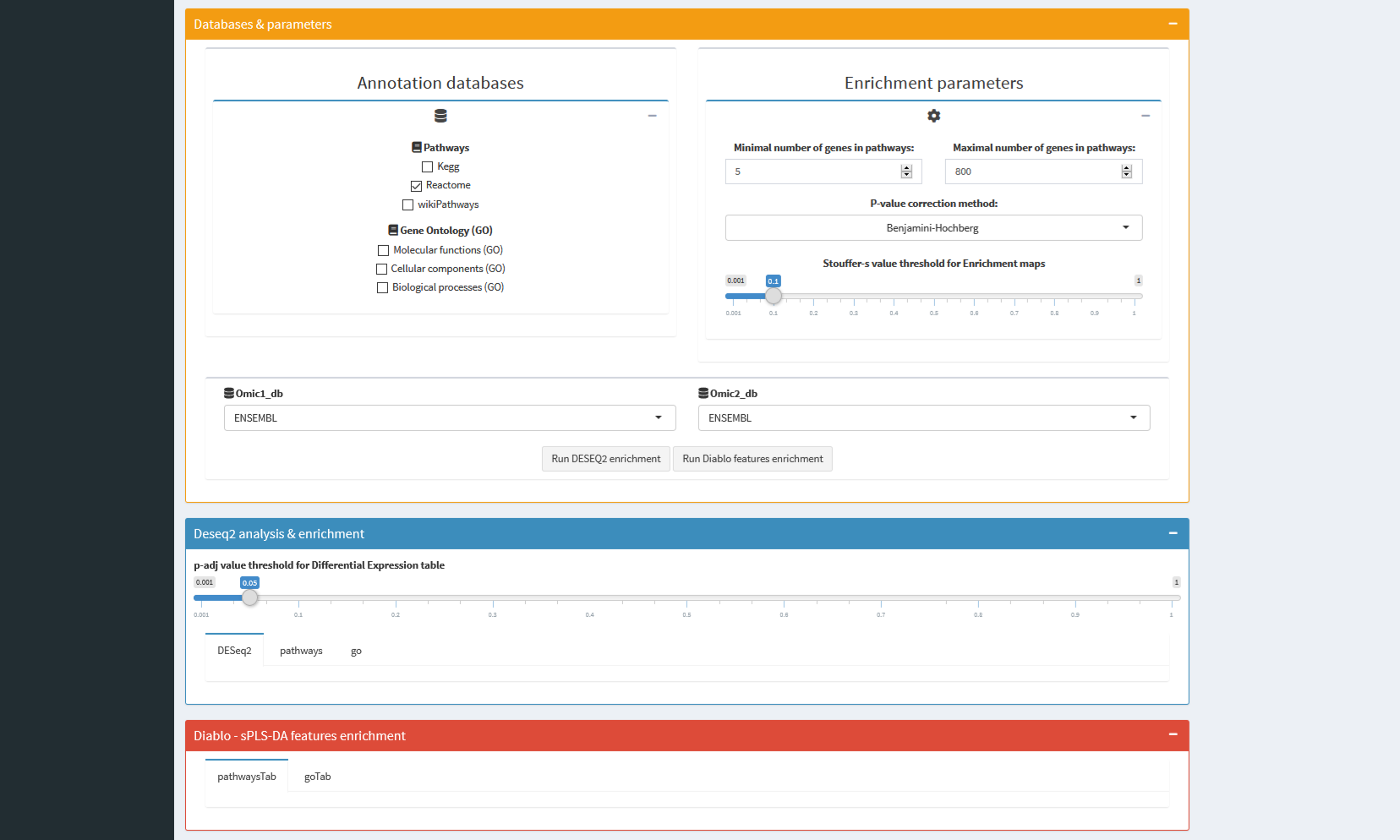 |
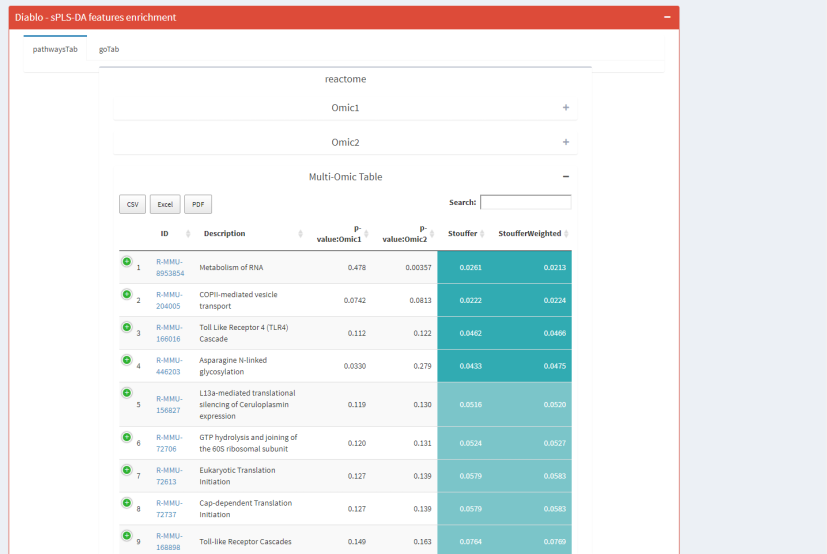 |
 |
👉 Some clicks (from 4 to number of PubMed queries)
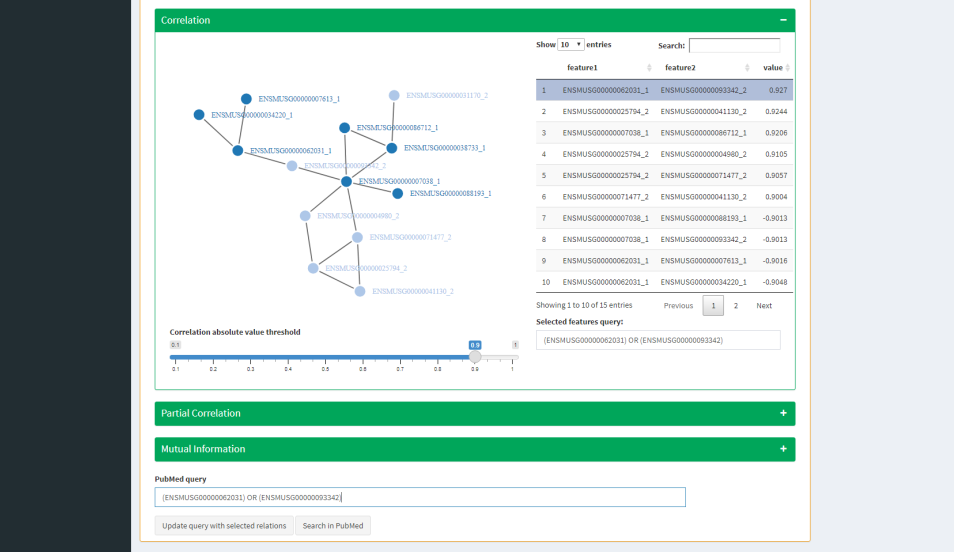
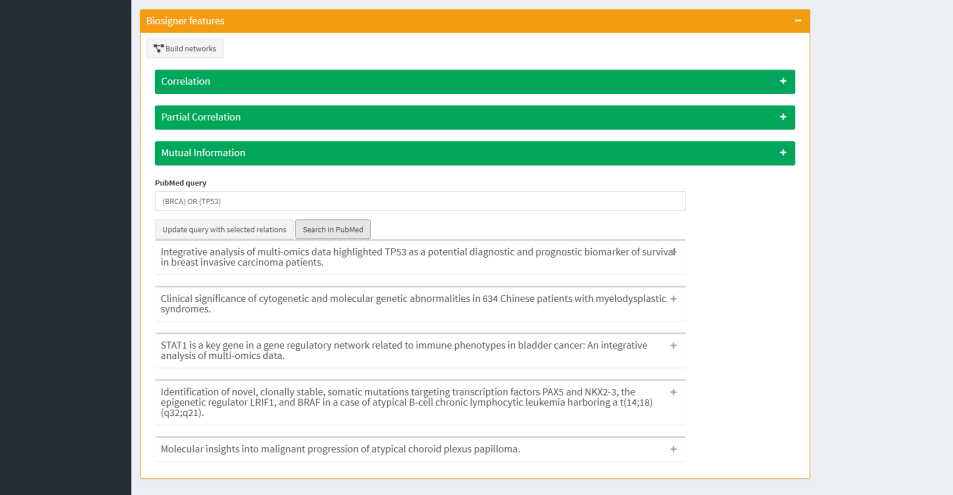
Assumption tab aims to help biological hypothesis making by network inference from feature relationship values (e.g correlation, partial correlation) and by a PubMed module.
You can find both functions:
- To compute network inference and to reveal feature relationships.
- To get PubMed articles based on your personalized query without leaving app.
| Network Inference | PubMed query |
|---|---|
 |
 |
You could retrieve different results computed by multiSight in Home tab by:
- Automatic report with all results in HTML and .doc documents.
- .RData with all results obtained by the graphical application.
Note that tables could be downloaded in a separated way in relative tabs.
MODELS: classification models you can use on future data.
DESeq2: differential expression analysis tables.
BIOSIGNATURES: DESeq2 tables thresholding and DIABLO multi-omics features selection method
Functional ENRICHMENTS: 6 databases functional enrichment for all omic datasets you provide enriched by Stouffer’s pooling p-value method giving a multi-omics enrichmentt able easily to discuss.
NETWORKS: network inference analysis with all features selected from all omic datasets according to DESeq2 tables thresholding or multi-omics feature selection (correlation, partial correlation, mutual information).
BIBLIOGRAPHY : a subset of PubMed articles relative to relations you choose in network inference tab.


