An Individual-Based Model (IBM) written in R.
This model simulates plant populations over time in order to try and understand the prevalance of genome duplication (polyploidy) that we see in flowering plants (angiosperms). In particular it focuses on one specific question:
How does disturbance on a landscape affect the establishment of new polyploid plant species?
By simulating, genetically explicit individuals (plants), on a landscape which can be disturbed, this model can be used to test which of the core benefits and costs to polyploidy most effect their establishment patterns in disturbed versus undisturbed environments.
From your R console:
install.packages("devtools")
library(devtools)
install_github("rosemckeon/ploidy")
library(disturploidy)From your R console/script running disturploidy() without parameters will run a quick simulations (ten generations) where no whole-genome duplication/distrubance occurs. You can turn these on with ploidy_prob and disturbance_freq respectively. By default polyploids that arise will incurr no costs/benefits from this mutation. See ?disturploidy for more details on model parameters which can be used to tailor these values, extend generations and alter the output format.
The model requires Devtools, Tidyverse and Tictoc to run. They will be installed with disturploidy, but not loaded. Make sure to load them:
library(devtools)
library(tidyverse)
library(tictoc)
You can also set simulations running on a server without entering an R console. Running simulations with BASH commands means you can automate processes to continuously run simulations if you so desire. For more details see here.
By default, model data is stored to an environment object (every generation) which can be loaded on completion by doing data(dploidy) from your R console. This file is overwritten everytime you run disturploidy() but you can specify a filename and/or logfilename to store information more permanently. Alternatively you can set return = T to output the data straight to console or assign it to your own objects.
Every generation there is:
- Survival of juveniles and adults.
- Germination followed by possible seed survival.
- Growth of juveniles and adults.
- Competition between adults for resources.
- Reproduction of adults and seed dispersal.
The life cycle of the plants in this model can be tailored to suit various life history strategies.
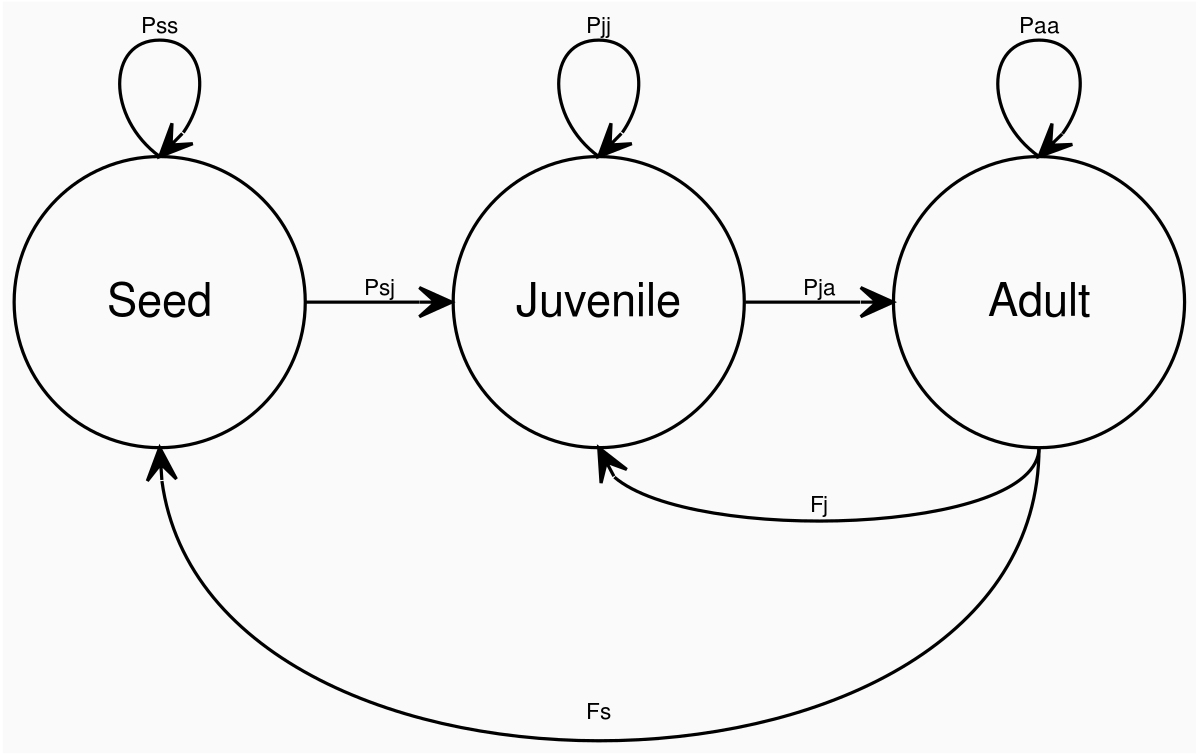
Figure 1: Disturploidy Life Cycle Graph. Individual plants in this model are represented by three life stages (seeds, juveniles and adults). Between generations, all individuals can remain at their current life stage or transition to the next. There is no back transitioning capability built into the model, so herbivory for instance is not considered.
Pss = probability of failing to germinate * chance of landing where there are adults * probability of seed survival
Seeds may fail to germinate due to chance, or because they land where adults are established.
Parameters: germination_prob, seed_survival_prob
Psj = chance of landing where there are no adults * probability of germination
Parameter: germination_prob
Pjj = probability of denisty-independant juvenile survival * chance of not growing to adult size * probability of not being outcompeted by adults in the same location
Density-independant juvenile survial is a parameter which is passed directly to the model using juvenile_selection_constant which assigns probabilities based on current size (the largest juveniles have the bast chance of survival). Density-independant mortality occurs between generations, over the winter period, before germination and growth. Density-dependant mortality occurs when adults become established and outcompete juveniles in the same location. Growth rates are calcualted based on alleles in the genomes of individuals and polyploids can be given a growth benefit to simulate the "gigas effects" of genome duplication (see genetics).
Parameters: growth_rate_loci, max_growth_rate, ploidy_growth_benefit, adult_size, juvenile_selection_constant.
Pja = chance of growing to adult size * probability of surviving competition
After the growing period, carrying capacity controls how many adults can survive on each landscape cell. The largest adults have the highest probabilities of surviving density-dependent mortallity during competition.
Parameters: growth_rate_loci, max_growth_rate, ploidy_growth_benefit, adult_size, carrying_capacity.
Paa = probability of density-independant adult survival * probability of surviving density-dependant competition
Adult survival probability is a set parameter passed to the model, which can be optionally reduced according to the occurance of inbreeding (see genetics). As above, carrying capacity controls how many adults can survive on each landscape cell, and the largest adults have the highest probabilities of surviving competition. Once established, new germinations do not take place in this location until the adult dies. This means the winning adult may survive for many generations, uncontested, depending on the value of adult_survival_prob.
Parameters: carrying_capacity, adult_survival_prob, inbreeding_locus, inbreeding_cost.
F = number of ovules * probability of fertilisation
The probability of fertilisation can be modified according to various mating situations:
- When gametes with uneven ploidy levels meet (ie: a haploid gamete and a diploid gamete).
- When a polyploid is selfing.
- When a diploid is selfing.
- When the ovules belong to a triploid.
Modifications occur sequentially, so for example: an increased fertilisaiton probability due to polyploid selfing may be overidden by a decreased fertilisation probability of having ovules that belong to a triploid.
Parameters: fertilisation_prob, uneven_matching_prob, selfing_polyploid_prob, selfing_diploid_prob, triploid_mum_prob.
Genomes of individuals contain alleles that are random numbers (to 5 decimal places) from a uniform distribution between 0 and 100. These alleles are used to confer two separate traits to individuals.
- Growth rate
- Inbreeding induced mortality
The amount of benefit gained for these traits by polyploids can be controlled via the parameters ploidy_growth_benefit and inbreeding_cost respectively. While the growth benefit directly controls the proportion of additional growth rate alleles polyploids are allowed to contribute, inbreeding sensitivity works differently. Inbreeding alleles are checked for homozygosity as a measure of being inbred or not. Polyploids are naturally more resillient to homozygosity of alleles. For this trait inbreeding_cost controls how much the mortality probability of inbred individuals should be increased.
Alleles are sampled (with replacement) from each loci of the parent genome to create the genomes of their gametes. pollen_range controls how freely alleles can travel across the landscape.
The landscape is always populated by diploid seeds. Under usual circumstances, as adults, these individuals will produce haploid gametes and generate more diploid seeds. However, genome duplication events within the reproductive cycle can cause polyploid indivuals to arise within the population.
Genome duplication occurs in independent gametes (simulating unreduced gametes) as well as in paired occurances (simulating nondisjuntion in early embryos). Now, instead of sampling alleles from each locus the entire parent genome is copied to it's gamete. The parameter ploidy_prob can be maniplulated to change the total rate of duplication events.
The ploidy level of this model is capped at four: tetraploids.
- Diploids produce haploid gametes unless duplication occurs, then they produce diploid gametes.
- Triploids produce 50/50 haploid/diploid gametes. When duplication occurs, a diploid gamete is garaunteed.
- Tetraploids always produce diploid gametes.
After ordinary gametogenesis and genome duplication have taken place, any of the alleles belonging to the new offspring may mutate. Mutation causes a new random number (to 5 decimal places from a uniform distribution between 0 and 100) to be chosen for that allele. The chance of identical alleles arising not by inheritance is, therefore, very slim. The parameter mutation_rate can be maniplulated to change the number of alleles that mutate in each generation.
Landscape boundaries wrap to produce a technically torus landscape with no edge.
Figure 2: Disturploidy Landscape. There is no edge.
Seeds disperse randomly in any direction, limited in distance from their maternal progenitor by seed_dispersal_range. When clonal_growth is set to TRUE, genets are formed and their ramets (juvenile forms genetically identical to the adult) are dispersed in a 1 cell radius of their progenitor.
Clonal growth requires some improvements - turning this parameter on is not currently recommended.
When disturbance occurs it increases the mortality of juveniles and adults during the usual survival period of the life-cycle. In this manner it could represent a natural disturbance such as a particularly harsh winter with extreme cold or floods. Or it could represent human disturbance like land clearance.
Parameters: disturbance_freq, disturbance_mortality_pob.
Simulations already run and the data collected are stored in a separate respository due to huge file size.
This project was funded by The Genetics Society as a Summer Studentship in 2019.