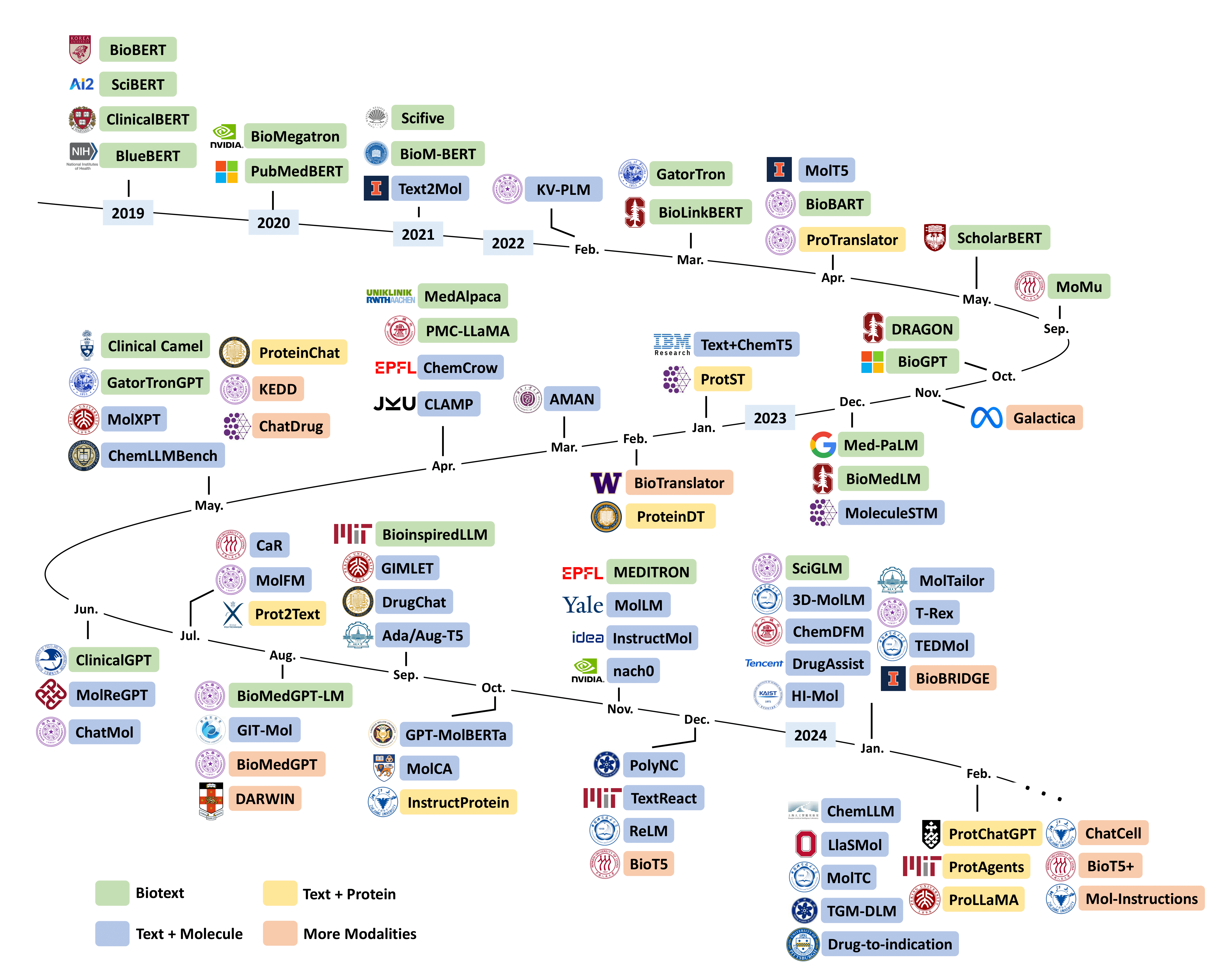
The repository for Leveraging Biomolecule and Natural Language through Multi-Modal Learning: A Survey, including related models, datasets/benchmarks, and other resource links.
🔥 We will keep this repository updated.
🌟 If you have a paper or resource you'd like to add, feel free to submit a pull request, open an issue, or email the author at qizhipei@ruc.edu.cn.
-
BioBERT: a pre-trained biomedical language representation model for biomedical text mining
-
SciBERT: A Pretrained Language Model for Scientific Text
-
(BlueBERT) Transfer Learning in Biomedical Natural Language Processing: An Evaluation of BERT and ELMo on Ten Benchmarking Datasets
-
Bio-Megatron: Larger Biomedical Domain Language Model
-
ClinicalBERT: Modeling Clinical Notes and Predicting Hospital Readmission
-
BioM-Transformers: Building Large Biomedical Language Models with BERT, ALBERT and ELECTRA
-
(PubMedBERT) Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing
-
SciFive: a text-to-text transformer model for biomedical literature
-
(DRAGON) Deep Bidirectional Language-Knowledge Graph Pretraining
-
LinkBERT: Pretraining Language Models with Document Links
-
BioBART: Pretraining and Evaluation of A Biomedical Generative Language Model
-
BioGPT: Generative Pre-trained Transformer for Biomedical Text Generation and Mining
-
GatorTron: A Large Clinical Language Model to Unlock Patient Information from Unstructured Electronic Health Records
-
Large language models encode clinical knowledge
-
(ScholarBERT) The Diminishing Returns of Masked Language Models to Science
-
PMC-LLaMA: Further Finetuning LLaMA on Medical Papers
-
BioMedGPT: Open Multimodal Generative Pre-trained Transformer for BioMedicine
-
(GatortronGPT) A study of generative large language model for medical research and healthcare
-
Clinical Camel: An Open-Source Expert-Level Medical Language Model with Dialogue-Based Knowledge Encoding
-
MEDITRON-70B: Scaling Medical Pretraining for Large Language Models
-
BioinspiredLLM: Conversational Large Language Model for the Mechanics of Biological and Bio-inspired Materials
-
ClinicalGPT: Large Language Models Finetuned with Diverse Medical Data and Comprehensive Evaluation
-
MedAlpaca - An Open-Source Collection of Medical Conversational AI Models and Training Data
-
SciGLM: Training Scientific Language Models with Self-Reflective Instruction Annotation and Tuning
-
BioMedLM: A 2.7B Parameter Language Model Trained On Biomedical Text
-
BioMistral: A Collection of Open-Source Pretrained Large Language Models for Medical Domains
-
Text2Mol: Cross-Modal Molecule Retrieval with Natural Language Queries
-
(MolT5) Translation between Molecules and Natural Language
-
(KV-PLM) A deep-learning system bridging molecule structure and biomedical text with comprehension comparable to human professionals
-
(MoMu) A Molecular Multimodal Foundation Model Associating Molecule Graphs with Natural Language
-
(Text+Chem T5) Unifying Molecular and Textual Representations via Multi-task Language Modelling
-
(CLAMP) Enhancing Activity Prediction Models in Drug Discovery with the Ability to Understand Human Language
-
GIMLET: A Unified Graph-Text Model for Instruction-Based Molecule Zero-Shot Learning
-
(HI-Mol) Data-Efficient Molecular Generation with Hierarchical Textual Inversion
-
MoleculeGPT: Instruction Following Large Language Models for Molecular Property Prediction
-
(ChemLLMBench) What indeed can GPT models do in chemistry? A comprehensive benchmark on eight tasks
-
MolXPT: Wrapping Molecules with Text for Generative Pre-training
-
(TextReact) Predictive Chemistry Augmented with Text Retrieval
-
MolCA: Molecular Graph-Language Modeling with Cross-Modal Projector and Uni-Modal Adapter
-
ReLM: Leveraging Language Models for Enhanced Chemical Reaction Prediction
-
(MoleculeSTM) Multi-modal Molecule Structure-text Model for Text-based Retrieval and Editing
-
(AMAN) Adversarial Modality Alignment Network for Cross-Modal Molecule Retrieval
-
MolLM: A Unified Language Model for Integrating Biomedical Text with 2D and 3D Molecular Representations
-
(MolReGPT) Empowering Molecule Discovery for Molecule-Caption Translation with Large Language Models: A ChatGPT Perspective
-
(CaR) Can Large Language Models Empower Molecular Property Prediction?
-
MolFM: A Multimodal Molecular Foundation Model
-
(ChatMol) Interactive Molecular Discovery with Natural Language
-
InstructMol: Multi-Modal Integration for Building a Versatile and Reliable Molecular Assistant in Drug Discovery
-
ChemCrow: Augmenting large-language models with chemistry tools
-
GPT-MolBERTa: GPT Molecular Features Language Model for molecular property prediction
-
nach0: Multimodal Natural and Chemical Languages Foundation Model
-
DrugChat: Towards Enabling ChatGPT-Like Capabilities on Drug Molecule Graphs
-
(Ada/Aug-T5) From Artificially Real to Real: Leveraging Pseudo Data from Large Language Models for Low-Resource Molecule Discovery
-
MolTailor: Tailoring Chemical Molecular Representation to Specific Tasks via Text Prompts
-
(TGM-DLM) Text-Guided Molecule Generation with Diffusion Language Model
-
GIT-Mol: A Multi-modal Large Language Model for Molecular Science with Graph, Image, and Text
-
PolyNC: a natural and chemical language model for the prediction of unified polymer properties
-
MolTC: Towards Molecular Relational Modeling In Language Models
-
T-Rex: Text-assisted Retrosynthesis Prediction
-
LlaSMol: Advancing Large Language Models for Chemistry with a Large-Scale, Comprehensive, High-Quality Instruction Tuning Dataset
-
(Drug-to-indication) Emerging Opportunities of Using Large Language Models for Translation Between Drug Molecules and Indications
-
ChemDFM: Dialogue Foundation Model for Chemistry
-
DrugAssist: A Large Language Model for Molecule Optimization
-
ChemLLM: A Chemical Large Language Model
-
(TEDMol) Text-guided Diffusion Model for 3D Molecule Generation
-
(3DToMolo) Sculpting Molecules in 3D: A Flexible Substructure Aware Framework for Text-Oriented Molecular Optimization
-
(ICMA) Large Language Models are In-Context Molecule Learners
-
Benchmarking Large Language Models for Molecule Prediction Tasks
-
DRAK: Unlocking Molecular Insights with Domain-Specific Retrieval-Augmented Knowledge in LLMs
-
3M-Diffusion: Latent Multi-Modal Diffusion for Text-Guided Generation of Molecular Graphs
-
(TSMMG) Instruction Multi-Constraint Molecular Generation Using a Teacher-Student Large Language Model
-
A Self-feedback Knowledge Elicitation Approach for Chemical Reaction Predictions
-
Atomas: Hierarchical Alignment on Molecule-Text for Unified Molecule Understanding and Generation
-
ReactXT: Understanding Molecular"Reaction-ship"via Reaction-Contextualized Molecule-Text Pretraining
-
LDMol: Text-Conditioned Molecule Diffusion Model Leveraging Chemically Informative Latent Space
-
(MV-Mol) Learning Multi-view Molecular Representations with Structured and Unstructured Knowledge
-
HIGHT: Hierarchical Graph Tokenization for Graph-Language Alignment
-
PRESTO: Progressive Pretraining Enhances Synthetic Chemistry Outcomes
-
3D-MolT5: Towards Unified 3D Molecule-Text Modeling with 3D Molecular Tokenization
-
MolecularGPT: Open Large Language Model (LLM) for Few-Shot Molecular Property Prediction
-
DrugLLM: Open Large Language Model for Few-shot Molecule Generation
-
(AMOLE) Vision Language Model is NOT All You Need: Augmentation Strategies for Molecule Language Models
-
Chemical Language Models Have Problems with Chemistry: A Case Study on Molecule Captioning Task
-
MolX: Enhancing Large Language Models for Molecular Learning with A Multi-Modal Extension
-
UniMoT: Unified Molecule-Text Language Model with Discrete Token Representation
-
OntoProtein: Protein Pretraining With Gene Ontology Embedding
-
ProTranslator: Zero-Shot Protein Function Prediction Using Textual Description
-
ProtST: Multi-Modality Learning of Protein Sequences and Biomedical Texts
-
InstructProtein: Aligning Human and Protein Language via Knowledge Instruction
-
(ProteinDT) A Text-guided Protein Design Framework
-
ProteinChat: Towards Achieving ChatGPT-Like Functionalities on Protein 3D Structures
-
Prot2Text: Multimodal Protein's Function Generation with GNNs and Transformers
-
ProtChatGPT: Towards Understanding Proteins with Large Language Models
-
ProtAgents: Protein discovery via large language model multi-agent collaborations combining physics and machine learning
-
ProLLaMA: A Protein Large Language Model for Multi-Task Protein Language Processing
-
ProtLLM: An Interleaved Protein-Language LLM with Protein-as-Word Pre-Training
-
ProtT3: Protein-to-Text Generation for Text-based Protein Understanding
-
ProteinCLIP: enhancing protein language models with natural language
-
ProLLM: Protein Chain-of-Thoughts Enhanced LLM for Protein-Protein Interaction Prediction
-
(PAAG) Functional Protein Design with Local Domain Alignment
-
(Pinal) Toward De Novo Protein Design from Natural Language
-
TourSynbio: A Multi-Modal Large Model and Agent Framework to Bridge Text and Protein Sequences for Protein Engineering
-
Galactica: A Large Language Model for Science
-
BioT5: Enriching Cross-modal Integration in Biology with Chemical Knowledge and Natural Language Associations
-
DARWIN Series: Domain Specific Large Language Models for Natural Science
-
BioMedGPT: Open Multimodal Generative Pre-trained Transformer for BioMedicine
-
(StructChem) Structured Chemistry Reasoning with Large Language Models
-
(BioTranslator) Multilingual translation for zero-shot biomedical classification using BioTranslator
-
Mol-Instructions: A Large-Scale Biomolecular Instruction Dataset for Large Language Models
-
(ChatDrug) ChatGPT-powered Conversational Drug Editing Using Retrieval and Domain Feedback
-
BioBridge: Bridging Biomedical Foundation Models via Knowledge Graphs
-
(KEDD) Towards Unified AI Drug Discovery with Multiple Knowledge Modalities
-
(Otter Knowledge) Knowledge Enhanced Representation Learning for Drug Discovery
-
ChatCell: Facilitating Single-Cell Analysis with Natural Language
-
LangCell: Language-Cell Pre-training for Cell Identity Understanding
-
BioT5+: Towards Generalized Biological Understanding with IUPAC Integration and Multi-task Tuning
-
MolBind: Multimodal Alignment of Language, Molecules, and Proteins
-
Uni-SMART: Universal Science Multimodal Analysis and Research Transformer
-
Tag-LLM: Repurposing General-Purpose LLMs for Specialized Domains
-
An Evaluation of Large Language Models in Bioinformatics Research
-
SciMind: A Multimodal Mixture-of-Experts Model for Advancing Pharmaceutical Sciences
- A Comprehensive Survey of Scientific Large Language Models and Their Applications in Scientific Discovery Arxiv 2406
- Bridging Text and Molecule: A Survey on Multimodal Frameworks for Molecule Arxiv 2403
- Bioinformatics and Biomedical Informatics with ChatGPT: Year One Review Arxiv 2403
- From Words to Molecules: A Survey of Large Language Models in Chemistry Arxiv 2402
- Scientific Language Modeling: A Quantitative Review of Large Language Models in Molecular Science Arxiv 2402
- Progress and Opportunities of Foundation Models in Bioinformatics Arxiv 2402
- Scientific Large Language Models: A Survey on Biological & Chemical Domains Arxiv 2401
- The Impact of Large Language Models on Scientific Discovery: a Preliminary Study using GPT-4 Arxiv 2311
- Transformers and Large Language Models for Chemistry and Drug Discovery Arxiv 2310
- Language models in molecular discovery Arxiv 2309
- What can Large Language Models do in chemistry? A comprehensive benchmark on eight tasks NeurIPS 2309
- Do Large Language Models Understand Chemistry? A Conversation with ChatGPT JCIM 2303
- A Systematic Survey of Chemical Pre-trained Models IJCAI 2023
- LLM4ScientificDiscovery
- SLM4Mol
- Scientific-LLM-Survey
- Awesome-Bio-Foundation-Models
- Awesome-Molecule-Text
- LLM4Mol
- Awesome-Chemical-Pre-trained-Models
- Awesome-Chemistry-Datasets
- Awesome-Docking
This repository is contributed and updated by QizhiPei and Lijun Wu. If you have questions, don't hesitate to open an issue or ask me via qizhipei@ruc.edu.cn or Lijun Wu via lijun_wu@outlook.com. We are happy to hear from you!
@article{pei2024leveraging,
title={Leveraging Biomolecule and Natural Language through Multi-Modal Learning: A Survey},
author={Pei, Qizhi and Wu, Lijun and Gao, Kaiyuan and Zhu, Jinhua and Wang, Yue and Wang, Zun and Qin, Tao and Yan, Rui},
journal={arXiv preprint arXiv:2403.01528},
year={2024}
}